Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
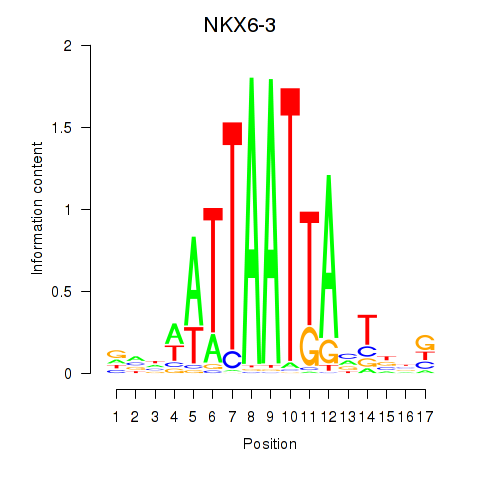
Results for NKX6-3
Z-value: 0.45
Transcription factors associated with NKX6-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX6-3
|
ENSG00000165066.13 | NK6 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX6-3 | hg38_v1_chr8_-_41650808_41650822 | -0.26 | 1.6e-01 | Click! |
Activity profile of NKX6-3 motif
Sorted Z-values of NKX6-3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_13495984 | 1.62 |
ENST00000282091.6
|
PTH
|
parathyroid hormone |
| chr11_-_13496018 | 1.49 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr9_+_122371014 | 1.44 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr9_+_122370523 | 1.38 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr9_+_122371036 | 1.38 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr8_+_91249307 | 1.16 |
ENST00000309536.6
ENST00000276609.8 |
SLC26A7
|
solute carrier family 26 member 7 |
| chr13_+_53028806 | 1.12 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr6_+_130018565 | 0.92 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chrX_+_108045050 | 0.63 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr18_+_22914115 | 0.55 |
ENST00000579124.5
ENST00000577588.5 ENST00000582354.5 ENST00000581819.5 |
RBBP8
|
RB binding protein 8, endonuclease |
| chrX_+_108044967 | 0.54 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr6_+_113857333 | 0.49 |
ENST00000612661.2
|
MARCKS
|
myristoylated alanine rich protein kinase C substrate |
| chr1_+_160400543 | 0.48 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr3_+_138621207 | 0.43 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr19_+_49513353 | 0.43 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr7_+_130344810 | 0.39 |
ENST00000497503.5
ENST00000463587.5 ENST00000461828.5 ENST00000474905.6 ENST00000494311.1 ENST00000466363.6 |
CPA5
|
carboxypeptidase A5 |
| chr5_-_143400716 | 0.37 |
ENST00000424646.6
ENST00000652686.1 |
NR3C1
|
nuclear receptor subfamily 3 group C member 1 |
| chr20_-_35147285 | 0.36 |
ENST00000374491.3
ENST00000374492.8 |
EDEM2
|
ER degradation enhancing alpha-mannosidase like protein 2 |
| chr3_+_136930469 | 0.35 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr17_-_40799939 | 0.34 |
ENST00000306658.8
|
KRT28
|
keratin 28 |
| chr3_+_138621225 | 0.33 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr6_-_49964160 | 0.32 |
ENST00000322066.4
|
DEFB114
|
defensin beta 114 |
| chr2_+_233917371 | 0.30 |
ENST00000324695.9
ENST00000433712.6 |
TRPM8
|
transient receptor potential cation channel subfamily M member 8 |
| chr16_+_86566821 | 0.29 |
ENST00000649859.1
|
FOXC2
|
forkhead box C2 |
| chr3_-_142029108 | 0.28 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr17_-_10469558 | 0.27 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4 |
| chr1_-_113871665 | 0.26 |
ENST00000528414.5
ENST00000460620.5 ENST00000359785.10 ENST00000420377.6 ENST00000525799.1 ENST00000538253.5 |
PTPN22
|
protein tyrosine phosphatase non-receptor type 22 |
| chr3_-_132684685 | 0.26 |
ENST00000512094.5
ENST00000632629.1 |
NPHP3
NPHP3-ACAD11
|
nephrocystin 3 NPHP3-ACAD11 readthrough (NMD candidate) |
| chr16_-_67483541 | 0.22 |
ENST00000290953.3
|
AGRP
|
agouti related neuropeptide |
| chr14_+_67364849 | 0.22 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2 subunit alpha |
| chr15_+_90191913 | 0.22 |
ENST00000559792.5
|
SEMA4B
|
semaphorin 4B |
| chr14_+_88385714 | 0.20 |
ENST00000045347.11
|
SPATA7
|
spermatogenesis associated 7 |
| chr14_+_88385643 | 0.19 |
ENST00000393545.9
ENST00000356583.9 ENST00000555401.5 ENST00000553885.5 |
SPATA7
|
spermatogenesis associated 7 |
| chrX_+_43656289 | 0.16 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr3_-_142000353 | 0.14 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2 |
| chr20_-_31390483 | 0.13 |
ENST00000376315.2
|
DEFB119
|
defensin beta 119 |
| chr15_+_92900189 | 0.11 |
ENST00000626874.2
ENST00000627622.1 ENST00000629346.2 ENST00000628375.2 ENST00000420239.7 ENST00000394196.9 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr1_-_151459169 | 0.11 |
ENST00000368863.6
ENST00000409503.5 ENST00000491586.5 ENST00000533351.5 |
POGZ
|
pogo transposable element derived with ZNF domain |
| chr4_-_106368772 | 0.10 |
ENST00000638719.4
|
GIMD1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr5_-_58999885 | 0.10 |
ENST00000317118.12
|
PDE4D
|
phosphodiesterase 4D |
| chr11_-_115256891 | 0.08 |
ENST00000545094.5
|
CADM1
|
cell adhesion molecule 1 |
| chr19_+_4402615 | 0.07 |
ENST00000301280.10
|
CHAF1A
|
chromatin assembly factor 1 subunit A |
| chr4_-_122621011 | 0.04 |
ENST00000611104.2
ENST00000648588.1 |
IL21
|
interleukin 21 |
| chrX_+_107777733 | 0.04 |
ENST00000509000.3
|
NCBP2L
|
nuclear cap binding protein subunit 2 like |
| chr1_-_151459471 | 0.04 |
ENST00000271715.7
|
POGZ
|
pogo transposable element derived with ZNF domain |
| chr1_-_151459198 | 0.01 |
ENST00000531094.5
|
POGZ
|
pogo transposable element derived with ZNF domain |
| chr15_+_92900338 | 0.01 |
ENST00000625990.3
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr12_-_52367478 | 0.01 |
ENST00000257901.7
|
KRT85
|
keratin 85 |
| chr8_+_76681208 | 0.01 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX6-3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.2 | 4.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.4 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.6 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.5 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.4 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.1 | 0.3 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.3 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) positive regulation of vascular wound healing(GO:0035470) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 1.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 1.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.3 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 1.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.2 | 0.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.2 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) translation initiation ternary complex(GO:0044207) |
| 0.1 | 0.3 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 4.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 3.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 1.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.4 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 3.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |