Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
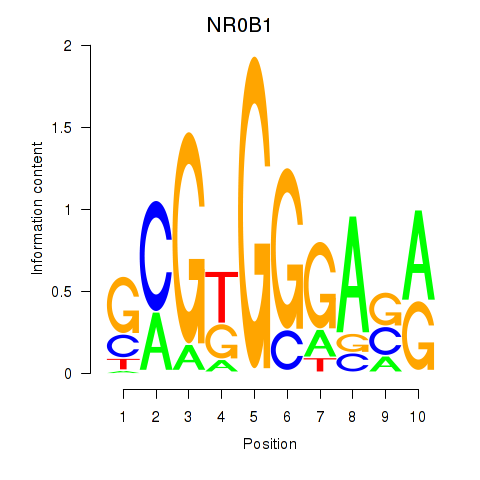
Results for NR0B1
Z-value: 0.85
Transcription factors associated with NR0B1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR0B1
|
ENSG00000169297.8 | nuclear receptor subfamily 0 group B member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR0B1 | hg38_v1_chrX_-_30309387_30309392 | -0.60 | 2.8e-04 | Click! |
Activity profile of NR0B1 motif
Sorted Z-values of NR0B1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_233636445 | 3.05 |
ENST00000344644.9
|
UGT1A10
|
UDP glucuronosyltransferase family 1 member A10 |
| chr4_+_42397473 | 2.60 |
ENST00000319234.5
|
SHISA3
|
shisa family member 3 |
| chr2_+_233636502 | 1.96 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase family 1 member A10 |
| chr1_-_46941464 | 1.90 |
ENST00000462347.5
ENST00000371905.1 ENST00000310638.9 |
CYP4A11
|
cytochrome P450 family 4 subfamily A member 11 |
| chr2_+_233671879 | 1.84 |
ENST00000354728.5
|
UGT1A9
|
UDP glucuronosyltransferase family 1 member A9 |
| chr2_+_42048012 | 1.58 |
ENST00000294964.6
|
PKDCC
|
protein kinase domain containing, cytoplasmic |
| chr3_+_111859796 | 1.44 |
ENST00000393925.7
|
PHLDB2
|
pleckstrin homology like domain family B member 2 |
| chr1_-_46941425 | 1.39 |
ENST00000371904.8
|
CYP4A11
|
cytochrome P450 family 4 subfamily A member 11 |
| chr17_-_7252054 | 1.26 |
ENST00000575783.5
ENST00000573600.5 |
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr15_-_64775574 | 1.22 |
ENST00000300069.5
|
RBPMS2
|
RNA binding protein, mRNA processing factor 2 |
| chr3_+_46883337 | 1.20 |
ENST00000313049.9
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr22_+_22895368 | 1.18 |
ENST00000390321.2
|
IGLC1
|
immunoglobulin lambda constant 1 |
| chr3_+_111859180 | 1.12 |
ENST00000412622.5
ENST00000431670.7 |
PHLDB2
|
pleckstrin homology like domain family B member 2 |
| chr6_-_100464912 | 1.06 |
ENST00000369208.8
|
SIM1
|
SIM bHLH transcription factor 1 |
| chr11_-_111912871 | 1.00 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B |
| chr6_+_29550407 | 0.97 |
ENST00000641137.1
|
OR2I1P
|
olfactory receptor family 2 subfamily I member 1 pseudogene |
| chrX_+_106920393 | 0.96 |
ENST00000336803.2
|
CLDN2
|
claudin 2 |
| chr1_-_93847150 | 0.95 |
ENST00000370244.5
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr1_+_31413806 | 0.94 |
ENST00000536384.2
|
SERINC2
|
serine incorporator 2 |
| chr3_+_111859736 | 0.91 |
ENST00000478922.1
ENST00000477695.5 |
PHLDB2
|
pleckstrin homology like domain family B member 2 |
| chr12_+_56080126 | 0.91 |
ENST00000411731.6
|
ERBB3
|
erb-b2 receptor tyrosine kinase 3 |
| chr10_-_21174187 | 0.90 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr17_+_78169127 | 0.88 |
ENST00000590201.1
|
SYNGR2
|
synaptogyrin 2 |
| chr17_-_68601357 | 0.81 |
ENST00000592554.2
|
FAM20A
|
FAM20A golgi associated secretory pathway pseudokinase |
| chr22_+_22668286 | 0.81 |
ENST00000390304.2
|
IGLV3-27
|
immunoglobulin lambda variable 3-27 |
| chr17_-_7251955 | 0.79 |
ENST00000318988.10
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr5_+_132369908 | 0.79 |
ENST00000435065.6
|
SLC22A5
|
solute carrier family 22 member 5 |
| chr17_+_47941562 | 0.79 |
ENST00000225573.5
ENST00000434554.7 ENST00000642017.2 |
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr3_+_111859284 | 0.79 |
ENST00000498699.5
|
PHLDB2
|
pleckstrin homology like domain family B member 2 |
| chr1_+_155277414 | 0.79 |
ENST00000368358.4
|
HCN3
|
hyperpolarization activated cyclic nucleotide gated potassium channel 3 |
| chr17_-_3696198 | 0.76 |
ENST00000345901.7
|
P2RX5
|
purinergic receptor P2X 5 |
| chr5_+_132369691 | 0.75 |
ENST00000245407.8
|
SLC22A5
|
solute carrier family 22 member 5 |
| chr19_-_14090695 | 0.73 |
ENST00000533683.7
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr17_-_3696133 | 0.72 |
ENST00000225328.10
|
P2RX5
|
purinergic receptor P2X 5 |
| chr7_-_520216 | 0.71 |
ENST00000405692.2
|
PDGFA
|
platelet derived growth factor subunit A |
| chr20_+_37179310 | 0.71 |
ENST00000373632.8
ENST00000237530.11 |
RPN2
|
ribophorin II |
| chr19_-_15934521 | 0.71 |
ENST00000402119.9
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11 |
| chrX_+_19343893 | 0.70 |
ENST00000540249.5
ENST00000545074.5 ENST00000379806.9 ENST00000423505.5 ENST00000422285.7 ENST00000417819.5 ENST00000355808.9 ENST00000379805.3 |
PDHA1
|
pyruvate dehydrogenase E1 subunit alpha 1 |
| chr20_+_37179109 | 0.70 |
ENST00000373622.9
|
RPN2
|
ribophorin II |
| chr1_-_147773341 | 0.70 |
ENST00000430508.1
ENST00000621517.1 |
GJA5
|
gap junction protein alpha 5 |
| chr7_+_90211686 | 0.67 |
ENST00000287908.7
ENST00000394621.7 ENST00000394626.5 |
STEAP2
|
STEAP2 metalloreductase |
| chr19_-_15934410 | 0.66 |
ENST00000326742.12
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11 |
| chr6_+_33428223 | 0.65 |
ENST00000682587.1
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
| chr10_+_73911104 | 0.65 |
ENST00000446342.5
ENST00000372764.4 |
PLAU
|
plasminogen activator, urokinase |
| chr7_-_120858303 | 0.64 |
ENST00000415871.5
ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr19_-_45406327 | 0.64 |
ENST00000593226.5
ENST00000418234.6 |
PPP1R13L
|
protein phosphatase 1 regulatory subunit 13 like |
| chr6_+_7541612 | 0.64 |
ENST00000418664.2
|
DSP
|
desmoplakin |
| chr2_+_47369467 | 0.62 |
ENST00000419334.1
|
EPCAM
|
epithelial cell adhesion molecule |
| chr6_-_93419545 | 0.62 |
ENST00000369297.1
ENST00000369303.9 ENST00000680224.1 ENST00000681532.1 ENST00000679565.1 |
EPHA7
|
EPH receptor A7 |
| chr17_+_57256514 | 0.62 |
ENST00000284073.7
ENST00000674964.1 |
MSI2
|
musashi RNA binding protein 2 |
| chr6_+_7541662 | 0.61 |
ENST00000379802.8
|
DSP
|
desmoplakin |
| chr8_-_42541578 | 0.59 |
ENST00000518384.1
|
SLC20A2
|
solute carrier family 20 member 2 |
| chr2_-_164841410 | 0.58 |
ENST00000342193.8
ENST00000375458.6 |
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr18_-_35344408 | 0.58 |
ENST00000261332.11
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chr12_+_56080155 | 0.58 |
ENST00000267101.8
|
ERBB3
|
erb-b2 receptor tyrosine kinase 3 |
| chr11_-_64745331 | 0.58 |
ENST00000377489.5
ENST00000354024.7 |
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr9_+_105694519 | 0.57 |
ENST00000374692.8
ENST00000434214.1 |
TMEM38B
|
transmembrane protein 38B |
| chr4_+_158672266 | 0.57 |
ENST00000684622.1
ENST00000683483.1 ENST00000684641.1 ENST00000682456.1 ENST00000684627.1 ENST00000511912.6 ENST00000684505.1 ENST00000683305.1 ENST00000684036.1 ENST00000683751.1 ENST00000684129.1 ENST00000307738.5 |
ETFDH
|
electron transfer flavoprotein dehydrogenase |
| chr1_+_25819926 | 0.57 |
ENST00000533762.5
ENST00000529116.5 ENST00000474295.5 ENST00000488327.6 ENST00000472643.5 ENST00000374303.7 ENST00000526894.5 ENST00000524618.5 ENST00000374307.9 |
MTFR1L
|
mitochondrial fission regulator 1 like |
| chr9_+_134326435 | 0.57 |
ENST00000481739.2
|
RXRA
|
retinoid X receptor alpha |
| chr2_+_26970628 | 0.56 |
ENST00000233121.7
ENST00000405074.7 |
MAPRE3
|
microtubule associated protein RP/EB family member 3 |
| chr6_+_17393576 | 0.56 |
ENST00000229922.7
ENST00000611958.4 |
CAP2
|
cyclase associated actin cytoskeleton regulatory protein 2 |
| chr17_-_3696033 | 0.55 |
ENST00000551178.5
ENST00000552276.5 ENST00000547178.5 |
P2RX5
|
purinergic receptor P2X 5 |
| chr21_+_38809596 | 0.54 |
ENST00000456966.1
|
ETS2
|
ETS proto-oncogene 2, transcription factor |
| chr1_-_23484171 | 0.54 |
ENST00000336689.8
ENST00000437606.6 |
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr14_-_106803221 | 0.53 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr10_+_122112957 | 0.51 |
ENST00000369001.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr13_-_33285682 | 0.51 |
ENST00000336934.10
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr1_+_61742418 | 0.51 |
ENST00000316485.11
ENST00000371158.6 ENST00000642238.2 ENST00000613764.4 |
PATJ
|
PATJ crumbs cell polarity complex component |
| chr19_-_49119092 | 0.50 |
ENST00000408991.4
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr5_-_42811884 | 0.50 |
ENST00000514985.6
ENST00000511224.5 ENST00000507920.5 ENST00000510965.1 |
SELENOP
|
selenoprotein P |
| chr16_+_1153202 | 0.50 |
ENST00000358590.8
ENST00000638323.1 |
CACNA1H
|
calcium voltage-gated channel subunit alpha1 H |
| chr20_+_58891981 | 0.49 |
ENST00000488652.6
ENST00000476935.6 ENST00000492907.6 ENST00000603546.2 |
GNAS
|
GNAS complex locus |
| chr7_+_149714769 | 0.49 |
ENST00000486744.3
|
KRBA1
|
KRAB-A domain containing 1 |
| chr17_+_7252024 | 0.49 |
ENST00000573513.5
ENST00000354429.6 ENST00000574255.5 ENST00000356683.6 ENST00000396627.6 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr1_+_25820146 | 0.49 |
ENST00000525713.5
ENST00000374301.7 |
MTFR1L
|
mitochondrial fission regulator 1 like |
| chr17_-_7252456 | 0.48 |
ENST00000571409.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr12_+_56079843 | 0.48 |
ENST00000549282.5
ENST00000549061.5 ENST00000683059.1 |
ERBB3
|
erb-b2 receptor tyrosine kinase 3 |
| chr17_+_47941506 | 0.48 |
ENST00000583599.6
|
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr3_+_72996842 | 0.45 |
ENST00000495566.1
|
PPP4R2
|
protein phosphatase 4 regulatory subunit 2 |
| chr4_-_176792913 | 0.44 |
ENST00000618562.2
|
VEGFC
|
vascular endothelial growth factor C |
| chr8_+_26291758 | 0.44 |
ENST00000522535.5
ENST00000665949.1 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr1_+_32741779 | 0.44 |
ENST00000401073.7
|
KIAA1522
|
KIAA1522 |
| chr11_-_1763894 | 0.43 |
ENST00000637915.1
ENST00000637815.2 ENST00000236671.7 ENST00000636571.1 ENST00000438213.6 ENST00000637387.1 ENST00000636843.1 ENST00000636397.1 ENST00000636615.1 |
CTSD
ENSG00000250644.3
|
cathepsin D novel protein |
| chr15_+_44537085 | 0.43 |
ENST00000424492.7
|
EIF3J
|
eukaryotic translation initiation factor 3 subunit J |
| chr8_+_64580357 | 0.43 |
ENST00000321870.3
|
BHLHE22
|
basic helix-loop-helix family member e22 |
| chrX_+_47585212 | 0.43 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr10_-_13707536 | 0.43 |
ENST00000632570.1
ENST00000477221.2 |
FRMD4A
|
FERM domain containing 4A |
| chr15_-_50765656 | 0.43 |
ENST00000261854.10
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr7_-_50782853 | 0.42 |
ENST00000401949.6
|
GRB10
|
growth factor receptor bound protein 10 |
| chrX_-_129654946 | 0.42 |
ENST00000429967.3
|
APLN
|
apelin |
| chr2_+_72917489 | 0.42 |
ENST00000258106.11
|
EMX1
|
empty spiracles homeobox 1 |
| chr10_+_84194527 | 0.42 |
ENST00000623527.4
|
CDHR1
|
cadherin related family member 1 |
| chr6_+_31158518 | 0.42 |
ENST00000376255.4
ENST00000376257.8 |
TCF19
|
transcription factor 19 |
| chr20_-_3173516 | 0.41 |
ENST00000360342.7
ENST00000645462.1 ENST00000337576.7 |
LZTS3
|
leucine zipper tumor suppressor family member 3 |
| chr3_+_188154150 | 0.41 |
ENST00000617246.5
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr1_-_94927079 | 0.41 |
ENST00000370206.9
ENST00000394202.8 |
CNN3
|
calponin 3 |
| chrX_+_49171918 | 0.41 |
ENST00000376322.7
|
PLP2
|
proteolipid protein 2 |
| chr15_+_44537136 | 0.41 |
ENST00000261868.10
ENST00000535391.5 |
EIF3J
|
eukaryotic translation initiation factor 3 subunit J |
| chr10_-_17454582 | 0.40 |
ENST00000377602.5
|
ST8SIA6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr16_+_58464273 | 0.40 |
ENST00000567454.5
|
NDRG4
|
NDRG family member 4 |
| chr1_+_212285383 | 0.40 |
ENST00000261461.7
|
PPP2R5A
|
protein phosphatase 2 regulatory subunit B'alpha |
| chr12_-_57846686 | 0.40 |
ENST00000548823.1
ENST00000398073.7 |
CTDSP2
|
CTD small phosphatase 2 |
| chr8_+_94641074 | 0.39 |
ENST00000423620.6
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr3_+_23917170 | 0.39 |
ENST00000643707.1
|
RPL15
|
ribosomal protein L15 |
| chr19_-_50333504 | 0.39 |
ENST00000474951.1
|
KCNC3
|
potassium voltage-gated channel subfamily C member 3 |
| chr17_+_7252237 | 0.38 |
ENST00000570500.5
|
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr1_+_2556361 | 0.38 |
ENST00000355716.5
|
TNFRSF14
|
TNF receptor superfamily member 14 |
| chr15_-_78620964 | 0.38 |
ENST00000326828.6
|
CHRNA3
|
cholinergic receptor nicotinic alpha 3 subunit |
| chr17_+_70104991 | 0.38 |
ENST00000587698.5
ENST00000587892.1 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr17_+_7252268 | 0.37 |
ENST00000396628.6
ENST00000574993.5 ENST00000573657.5 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr5_-_58999885 | 0.37 |
ENST00000317118.12
|
PDE4D
|
phosphodiesterase 4D |
| chr1_-_196608359 | 0.37 |
ENST00000609185.5
ENST00000451324.6 ENST00000367433.9 ENST00000294725.14 |
KCNT2
|
potassium sodium-activated channel subfamily T member 2 |
| chr15_+_40844064 | 0.37 |
ENST00000568823.5
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr3_-_127590707 | 0.37 |
ENST00000296210.11
ENST00000355552.8 |
TPRA1
|
transmembrane protein adipocyte associated 1 |
| chr1_+_15617415 | 0.36 |
ENST00000480945.6
|
DDI2
|
DNA damage inducible 1 homolog 2 |
| chr19_+_8364146 | 0.36 |
ENST00000301455.7
ENST00000393962.6 |
ANGPTL4
|
angiopoietin like 4 |
| chr10_-_17617235 | 0.36 |
ENST00000466335.1
|
HACD1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr8_-_141000937 | 0.36 |
ENST00000520892.5
|
PTK2
|
protein tyrosine kinase 2 |
| chr11_-_117232492 | 0.36 |
ENST00000530269.1
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chr7_-_108456700 | 0.36 |
ENST00000418239.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr11_-_20160473 | 0.35 |
ENST00000524983.3
|
DBX1
|
developing brain homeobox 1 |
| chr1_+_153728042 | 0.34 |
ENST00000318967.7
ENST00000435409.6 |
INTS3
|
integrator complex subunit 3 |
| chr19_-_3573629 | 0.34 |
ENST00000592652.1
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr7_+_90211830 | 0.34 |
ENST00000394622.6
ENST00000394632.5 ENST00000426158.1 ENST00000402625.6 |
STEAP2
|
STEAP2 metalloreductase |
| chr6_-_10412879 | 0.34 |
ENST00000465858.1
|
TFAP2A
|
transcription factor AP-2 alpha |
| chr12_+_110280602 | 0.33 |
ENST00000552636.2
|
ATP2A2
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| chr4_-_113761927 | 0.33 |
ENST00000296402.9
|
CAMK2D
|
calcium/calmodulin dependent protein kinase II delta |
| chr15_+_40844506 | 0.33 |
ENST00000568580.5
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr16_+_9091593 | 0.33 |
ENST00000327827.12
|
C16orf72
|
chromosome 16 open reading frame 72 |
| chr11_+_64035925 | 0.33 |
ENST00000682287.1
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr6_+_17393657 | 0.33 |
ENST00000493172.5
ENST00000465994.5 |
CAP2
|
cyclase associated actin cytoskeleton regulatory protein 2 |
| chr13_-_37869723 | 0.32 |
ENST00000426868.6
ENST00000379705.8 ENST00000338947.9 ENST00000355779.6 ENST00000358477.6 ENST00000379673.2 |
TRPC4
|
transient receptor potential cation channel subfamily C member 4 |
| chr19_+_49157741 | 0.32 |
ENST00000598691.5
|
TRPM4
|
transient receptor potential cation channel subfamily M member 4 |
| chr17_-_7252482 | 0.32 |
ENST00000572043.5
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr17_-_3668640 | 0.32 |
ENST00000611779.4
|
TAX1BP3
|
Tax1 binding protein 3 |
| chr7_-_100896919 | 0.31 |
ENST00000445236.3
ENST00000441605.2 |
ACHE
|
acetylcholinesterase (Cartwright blood group) |
| chr19_-_4722691 | 0.31 |
ENST00000598360.5
|
DPP9
|
dipeptidyl peptidase 9 |
| chr8_-_101205455 | 0.31 |
ENST00000520984.5
|
ZNF706
|
zinc finger protein 706 |
| chr12_-_27780236 | 0.31 |
ENST00000381273.4
|
MANSC4
|
MANSC domain containing 4 |
| chr14_+_51651976 | 0.31 |
ENST00000554778.1
|
FRMD6
|
FERM domain containing 6 |
| chr3_-_45915596 | 0.31 |
ENST00000472635.5
ENST00000492333.5 |
LZTFL1
|
leucine zipper transcription factor like 1 |
| chr2_+_54115396 | 0.31 |
ENST00000406041.5
|
ACYP2
|
acylphosphatase 2 |
| chr11_-_119101814 | 0.31 |
ENST00000682791.1
ENST00000639704.1 ENST00000354202.9 |
DPAGT1
|
dolichyl-phosphate N-acetylglucosaminephosphotransferase 1 |
| chr10_-_77637558 | 0.31 |
ENST00000372421.10
ENST00000639370.1 ENST00000640773.1 ENST00000638895.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr3_+_172040554 | 0.30 |
ENST00000336824.8
ENST00000423424.5 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr2_+_85753984 | 0.30 |
ENST00000306279.4
|
ATOH8
|
atonal bHLH transcription factor 8 |
| chr6_+_17393607 | 0.30 |
ENST00000489374.5
ENST00000378990.6 |
CAP2
|
cyclase associated actin cytoskeleton regulatory protein 2 |
| chr11_+_67391975 | 0.30 |
ENST00000307980.7
|
RAD9A
|
RAD9 checkpoint clamp component A |
| chr12_+_106774630 | 0.30 |
ENST00000392839.6
ENST00000548914.5 ENST00000355478.6 ENST00000552619.1 ENST00000549643.5 ENST00000392837.9 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr22_-_23838987 | 0.29 |
ENST00000318109.12
ENST00000404056.1 ENST00000406855.7 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr15_+_40844171 | 0.29 |
ENST00000563656.5
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr19_-_2051224 | 0.29 |
ENST00000309340.11
ENST00000589534.2 ENST00000250896.9 ENST00000589509.5 |
MKNK2
|
MAPK interacting serine/threonine kinase 2 |
| chr16_+_4795357 | 0.29 |
ENST00000586005.6
|
SMIM22
|
small integral membrane protein 22 |
| chr20_+_297553 | 0.28 |
ENST00000500893.4
|
ZCCHC3
|
zinc finger CCHC-type containing 3 |
| chr6_+_143060853 | 0.28 |
ENST00000447498.5
ENST00000646199.1 ENST00000357847.9 ENST00000629020.2 ENST00000367596.5 ENST00000494282.6 ENST00000275235.8 |
AIG1
|
androgen induced 1 |
| chr20_+_58891738 | 0.28 |
ENST00000682803.1
|
GNAS
|
GNAS complex locus |
| chr16_+_2019777 | 0.28 |
ENST00000566435.4
|
NPW
|
neuropeptide W |
| chr5_+_176365455 | 0.28 |
ENST00000310389.6
|
ARL10
|
ADP ribosylation factor like GTPase 10 |
| chr5_+_93583212 | 0.28 |
ENST00000327111.8
|
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr20_+_3046008 | 0.28 |
ENST00000380325.4
|
MRPS26
|
mitochondrial ribosomal protein S26 |
| chr3_-_122993232 | 0.27 |
ENST00000650207.1
ENST00000616742.4 ENST00000393583.6 |
SEMA5B
|
semaphorin 5B |
| chr16_+_19168207 | 0.27 |
ENST00000355377.7
ENST00000568115.5 |
SYT17
|
synaptotagmin 17 |
| chrX_+_129980965 | 0.27 |
ENST00000607874.1
|
BCORL1
|
BCL6 corepressor like 1 |
| chr15_+_40844018 | 0.26 |
ENST00000344051.8
ENST00000562057.6 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr5_+_96702849 | 0.26 |
ENST00000508197.1
|
CAST
|
calpastatin |
| chr7_+_143263412 | 0.26 |
ENST00000409500.7
ENST00000443571.6 ENST00000358406.10 ENST00000479303.1 |
GSTK1
|
glutathione S-transferase kappa 1 |
| chr16_+_1153098 | 0.26 |
ENST00000348261.11
|
CACNA1H
|
calcium voltage-gated channel subunit alpha1 H |
| chr2_+_172084728 | 0.26 |
ENST00000361609.4
|
DLX1
|
distal-less homeobox 1 |
| chr8_+_22165140 | 0.26 |
ENST00000397814.7
ENST00000354870.5 |
BMP1
|
bone morphogenetic protein 1 |
| chr12_-_42238261 | 0.26 |
ENST00000380790.4
|
YAF2
|
YY1 associated factor 2 |
| chr19_+_49119531 | 0.26 |
ENST00000334186.9
|
PPFIA3
|
PTPRF interacting protein alpha 3 |
| chr14_-_106658251 | 0.26 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr12_-_6375556 | 0.26 |
ENST00000228916.7
|
SCNN1A
|
sodium channel epithelial 1 subunit alpha |
| chr4_-_2756288 | 0.26 |
ENST00000510267.5
ENST00000315423.12 ENST00000503235.1 |
TNIP2
|
TNFAIP3 interacting protein 2 |
| chr19_-_40348510 | 0.25 |
ENST00000582006.5
ENST00000582783.5 |
C19orf47
|
chromosome 19 open reading frame 47 |
| chr1_-_153986329 | 0.25 |
ENST00000368575.5
|
RAB13
|
RAB13, member RAS oncogene family |
| chr1_+_38991239 | 0.25 |
ENST00000432648.8
ENST00000446189.6 ENST00000372984.8 |
AKIRIN1
|
akirin 1 |
| chr10_+_84194621 | 0.25 |
ENST00000332904.7
|
CDHR1
|
cadherin related family member 1 |
| chr15_+_90903731 | 0.25 |
ENST00000559965.5
|
MAN2A2
|
mannosidase alpha class 2A member 2 |
| chr3_+_172039556 | 0.24 |
ENST00000415807.7
ENST00000421757.5 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr8_-_73746830 | 0.24 |
ENST00000524300.6
ENST00000523558.5 ENST00000521210.5 ENST00000355780.9 ENST00000524104.5 ENST00000521736.5 ENST00000521447.5 ENST00000517542.5 ENST00000521451.5 ENST00000521419.5 ENST00000518502.5 |
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr4_+_155758990 | 0.24 |
ENST00000505154.5
ENST00000652626.1 ENST00000502959.5 ENST00000264424.13 ENST00000505764.5 ENST00000507146.5 ENST00000503520.5 |
GUCY1B1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr12_-_42238323 | 0.24 |
ENST00000555248.2
|
YAF2
|
YY1 associated factor 2 |
| chr2_-_43595963 | 0.24 |
ENST00000405006.8
|
THADA
|
THADA armadillo repeat containing |
| chr2_+_48440757 | 0.24 |
ENST00000294952.13
ENST00000281394.8 ENST00000449090.6 |
PPP1R21
|
protein phosphatase 1 regulatory subunit 21 |
| chr12_+_52037232 | 0.24 |
ENST00000553200.1
|
NR4A1
|
nuclear receptor subfamily 4 group A member 1 |
| chrX_+_134796758 | 0.23 |
ENST00000414371.6
|
PABIR3
|
PABIR family member 3 |
| chr2_-_65432591 | 0.23 |
ENST00000356388.9
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr6_+_12749419 | 0.23 |
ENST00000406205.7
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr10_-_689613 | 0.23 |
ENST00000280886.12
ENST00000634311.1 |
DIP2C
|
disco interacting protein 2 homolog C |
| chr8_-_141001192 | 0.23 |
ENST00000521059.5
|
PTK2
|
protein tyrosine kinase 2 |
| chr3_-_184261547 | 0.22 |
ENST00000296238.4
|
CAMK2N2
|
calcium/calmodulin dependent protein kinase II inhibitor 2 |
| chr17_-_7207340 | 0.22 |
ENST00000650301.1
|
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr2_+_27078598 | 0.22 |
ENST00000380320.9
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr5_+_96702821 | 0.22 |
ENST00000675033.1
|
CAST
|
calpastatin |
| chr19_-_4722767 | 0.22 |
ENST00000600621.5
|
DPP9
|
dipeptidyl peptidase 9 |
| chr1_+_116909869 | 0.22 |
ENST00000393203.3
|
PTGFRN
|
prostaglandin F2 receptor inhibitor |
| chr16_-_1964803 | 0.21 |
ENST00000526522.5
ENST00000527302.1 ENST00000529806.5 ENST00000343262.9 ENST00000563194.1 |
RPS2
|
ribosomal protein S2 |
| chr19_-_40348375 | 0.21 |
ENST00000392035.6
ENST00000683109.1 |
C19orf47
|
chromosome 19 open reading frame 47 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR0B1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.8 | 6.9 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.5 | 1.5 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.5 | 1.4 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.3 | 4.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 0.7 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.2 | 0.7 | GO:0098905 | pulmonary valve formation(GO:0003193) foramen ovale closure(GO:0035922) regulation of bundle of His cell action potential(GO:0098905) |
| 0.2 | 1.3 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 0.2 | 0.8 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.2 | 0.6 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 | 0.6 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.2 | 1.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 1.6 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.2 | 0.7 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 0.9 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.4 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 1.0 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.4 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.1 | 0.3 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.1 | 0.4 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.3 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.1 | 3.0 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.5 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.8 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 1.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.3 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.3 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 1.5 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.3 | GO:0061075 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.7 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.3 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.4 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.8 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.5 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.3 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.9 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.3 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.1 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 2.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.2 | GO:1902811 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 | 0.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:1904000 | positive regulation of eating behavior(GO:1904000) regulation of small intestine smooth muscle contraction(GO:1904347) positive regulation of small intestine smooth muscle contraction(GO:1904349) small intestine smooth muscle contraction(GO:1990770) |
| 0.0 | 0.2 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.4 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 1.9 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 1.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 1.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.9 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.4 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.4 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.2 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.6 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0086017 | Purkinje myocyte action potential(GO:0086017) |
| 0.0 | 0.6 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.3 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 2.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.2 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 0.0 | 0.3 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.3 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.4 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.3 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.5 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.6 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.8 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 1.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:2000782 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.3 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 4.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.9 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.6 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 1.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.0 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.7 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.3 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 1.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 2.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 2.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.8 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 2.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.4 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.3 | 1.3 | GO:0004733 | pyridoxamine-phosphate oxidase activity(GO:0004733) |
| 0.2 | 1.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 1.5 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.2 | 0.6 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 0.7 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 2.0 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 1.0 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 7.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 0.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 2.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.7 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 1.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.4 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.1 | 0.3 | GO:0086039 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.1 | 1.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.8 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.2 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.1 | 0.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.9 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 1.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 1.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 3.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 1.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.6 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 2.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.9 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.8 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 2.2 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |