Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
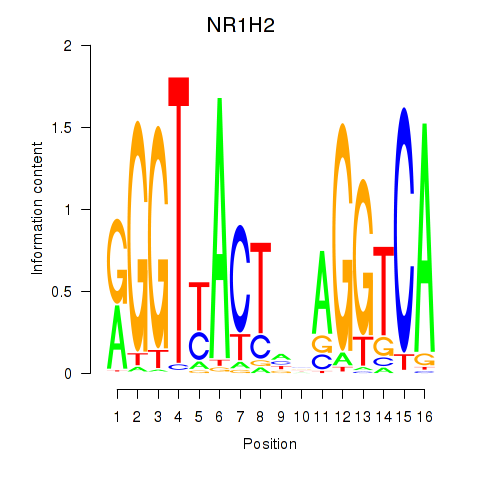
Results for NR1H2
Z-value: 0.88
Transcription factors associated with NR1H2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1H2
|
ENSG00000131408.15 | nuclear receptor subfamily 1 group H member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1H2 | hg38_v1_chr19_+_50358571_50358611 | -0.24 | 1.9e-01 | Click! |
Activity profile of NR1H2 motif
Sorted Z-values of NR1H2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_78063851 | 6.85 |
ENST00000281030.2
|
THRSP
|
thyroid hormone responsive |
| chr10_-_68527498 | 2.34 |
ENST00000609923.6
|
SLC25A16
|
solute carrier family 25 member 16 |
| chr2_-_216081759 | 2.05 |
ENST00000265322.8
|
PECR
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr12_+_31962466 | 2.02 |
ENST00000381054.3
|
RESF1
|
retroelement silencing factor 1 |
| chr4_-_70666884 | 1.70 |
ENST00000510614.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr4_-_70666961 | 1.64 |
ENST00000510437.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr9_-_104928139 | 1.56 |
ENST00000423487.6
ENST00000374733.1 ENST00000374736.8 ENST00000678995.1 |
ABCA1
|
ATP binding cassette subfamily A member 1 |
| chr17_-_17823593 | 1.48 |
ENST00000423161.3
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr19_+_15640880 | 1.47 |
ENST00000586182.6
ENST00000221307.13 ENST00000591058.5 |
CYP4F3
|
cytochrome P450 family 4 subfamily F member 3 |
| chr16_-_29745951 | 1.38 |
ENST00000329410.4
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr12_+_56118241 | 1.37 |
ENST00000551790.5
ENST00000552345.1 ENST00000257940.7 ENST00000551880.1 |
ESYT1
ZC3H10
|
extended synaptotagmin 1 zinc finger CCCH-type containing 10 |
| chr16_+_72008588 | 1.35 |
ENST00000572887.5
ENST00000219240.9 ENST00000574309.5 ENST00000576145.1 |
DHODH
|
dihydroorotate dehydrogenase (quinone) |
| chr19_+_17511835 | 1.31 |
ENST00000595782.1
|
PGLS
|
6-phosphogluconolactonase |
| chr19_+_15641280 | 1.30 |
ENST00000585846.1
|
CYP4F3
|
cytochrome P450 family 4 subfamily F member 3 |
| chrX_-_2929324 | 1.23 |
ENST00000381154.6
|
ARSD
|
arylsulfatase D |
| chr12_-_113403873 | 1.23 |
ENST00000552280.5
ENST00000257549.9 |
SDS
|
serine dehydratase |
| chr12_-_7018465 | 1.14 |
ENST00000261407.9
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3 |
| chr14_+_24130659 | 1.12 |
ENST00000267426.6
|
FITM1
|
fat storage inducing transmembrane protein 1 |
| chr19_-_2042066 | 1.11 |
ENST00000591588.1
ENST00000591142.5 |
MKNK2
|
MAPK interacting serine/threonine kinase 2 |
| chr8_-_28083920 | 1.09 |
ENST00000413272.7
|
NUGGC
|
nuclear GTPase, germinal center associated |
| chr4_-_2963327 | 1.08 |
ENST00000398071.4
ENST00000502735.5 ENST00000314262.10 ENST00000416614.7 |
NOP14
|
NOP14 nucleolar protein |
| chr11_+_87037956 | 1.03 |
ENST00000525018.5
|
TMEM135
|
transmembrane protein 135 |
| chr4_+_107989714 | 1.03 |
ENST00000505878.4
ENST00000603302.5 ENST00000638621.1 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr19_-_38315749 | 1.01 |
ENST00000589247.1
ENST00000329420.12 ENST00000591784.5 |
YIF1B
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chr4_-_83284752 | 1.01 |
ENST00000311461.7
ENST00000647002.2 ENST00000311469.9 |
COQ2
|
coenzyme Q2, polyprenyltransferase |
| chr11_+_32829903 | 0.99 |
ENST00000257836.4
|
PRRG4
|
proline rich and Gla domain 4 |
| chr17_-_3691748 | 0.97 |
ENST00000552723.1
|
P2RX5
|
purinergic receptor P2X 5 |
| chr11_+_87037915 | 0.96 |
ENST00000526733.5
ENST00000305494.6 ENST00000532959.5 |
TMEM135
|
transmembrane protein 135 |
| chr11_-_118212885 | 0.95 |
ENST00000524477.5
|
JAML
|
junction adhesion molecule like |
| chr19_+_40586774 | 0.93 |
ENST00000594298.5
ENST00000597396.5 |
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr7_+_80624071 | 0.92 |
ENST00000438020.5
|
CD36
|
CD36 molecule |
| chr17_-_28368012 | 0.91 |
ENST00000555059.2
|
ENSG00000273171.1
|
novel protein, readthrough between VTN and SEBOX |
| chr12_-_46832370 | 0.90 |
ENST00000546940.1
|
SLC38A4
|
solute carrier family 38 member 4 |
| chr16_-_68000549 | 0.90 |
ENST00000575510.5
|
DPEP2
|
dipeptidase 2 |
| chr14_+_104579757 | 0.90 |
ENST00000331952.6
ENST00000557649.6 |
C14orf180
|
chromosome 14 open reading frame 180 |
| chr1_-_161238196 | 0.90 |
ENST00000367983.9
ENST00000506209.5 ENST00000367980.6 ENST00000628566.2 |
NR1I3
|
nuclear receptor subfamily 1 group I member 3 |
| chr19_-_38315900 | 0.90 |
ENST00000592694.5
|
YIF1B
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chr10_-_70888546 | 0.89 |
ENST00000299299.4
|
PCBD1
|
pterin-4 alpha-carbinolamine dehydratase 1 |
| chr15_+_81296913 | 0.89 |
ENST00000394652.6
|
IL16
|
interleukin 16 |
| chr1_-_237004440 | 0.87 |
ENST00000464121.3
|
MT1HL1
|
metallothionein 1H like 1 |
| chr12_+_56118396 | 0.86 |
ENST00000546903.1
|
ZC3H10
|
zinc finger CCCH-type containing 10 |
| chr1_+_44808482 | 0.85 |
ENST00000450269.5
ENST00000409335.6 |
BTBD19
|
BTB domain containing 19 |
| chr16_+_527698 | 0.84 |
ENST00000219611.7
ENST00000562370.5 ENST00000568988.5 |
CAPN15
|
calpain 15 |
| chr19_-_6670151 | 0.83 |
ENST00000675206.1
|
TNFSF14
|
TNF superfamily member 14 |
| chr11_+_119168705 | 0.82 |
ENST00000409109.6
ENST00000409991.5 ENST00000292199.6 |
NLRX1
|
NLR family member X1 |
| chr11_+_119168188 | 0.81 |
ENST00000454811.5
ENST00000409265.8 ENST00000449394.5 |
NLRX1
|
NLR family member X1 |
| chr16_-_3577375 | 0.78 |
ENST00000359128.10
|
NLRC3
|
NLR family CARD domain containing 3 |
| chr16_-_3577288 | 0.78 |
ENST00000324659.12
|
NLRC3
|
NLR family CARD domain containing 3 |
| chr19_-_6670117 | 0.77 |
ENST00000245912.7
|
TNFSF14
|
TNF superfamily member 14 |
| chr19_+_17511606 | 0.76 |
ENST00000252603.7
ENST00000600923.5 |
PGLS
|
6-phosphogluconolactonase |
| chr19_-_38315919 | 0.76 |
ENST00000591755.5
ENST00000337679.12 ENST00000339413.11 |
YIF1B
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chr20_+_6007245 | 0.76 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr17_+_4771878 | 0.76 |
ENST00000270560.4
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr14_-_73996219 | 0.75 |
ENST00000553284.5
|
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5 (inactive) |
| chr2_+_233195433 | 0.75 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr1_-_54623518 | 0.74 |
ENST00000302250.7
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151 member A |
| chr17_+_79022814 | 0.74 |
ENST00000578229.5
|
C1QTNF1
|
C1q and TNF related 1 |
| chr4_-_39032343 | 0.73 |
ENST00000381938.4
|
TMEM156
|
transmembrane protein 156 |
| chr19_-_35900532 | 0.72 |
ENST00000396901.5
ENST00000641389.2 ENST00000585925.7 |
NFKBID
|
NFKB inhibitor delta |
| chr1_-_161238223 | 0.72 |
ENST00000515452.1
|
NR1I3
|
nuclear receptor subfamily 1 group I member 3 |
| chrX_+_48683763 | 0.70 |
ENST00000376701.5
|
WAS
|
WASP actin nucleation promoting factor |
| chr15_-_88467353 | 0.68 |
ENST00000312475.5
ENST00000558531.1 |
MRPL46
|
mitochondrial ribosomal protein L46 |
| chr1_-_161238163 | 0.68 |
ENST00000367982.8
|
NR1I3
|
nuclear receptor subfamily 1 group I member 3 |
| chr15_-_60391127 | 0.68 |
ENST00000559350.5
ENST00000558986.5 ENST00000560389.5 |
ANXA2
|
annexin A2 |
| chr17_-_8248035 | 0.68 |
ENST00000651323.1
|
CTC1
|
CST telomere replication complex component 1 |
| chr19_+_16143678 | 0.68 |
ENST00000613986.4
ENST00000593031.1 |
HSH2D
|
hematopoietic SH2 domain containing |
| chr2_-_164841410 | 0.68 |
ENST00000342193.8
ENST00000375458.6 |
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr17_-_17577440 | 0.68 |
ENST00000395782.5
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr11_+_35186820 | 0.68 |
ENST00000531110.6
ENST00000525685.6 |
CD44
|
CD44 molecule (Indian blood group) |
| chr16_-_11629210 | 0.68 |
ENST00000576036.5
|
LITAF
|
lipopolysaccharide induced TNF factor |
| chr16_+_30182969 | 0.67 |
ENST00000561815.5
|
CORO1A
|
coronin 1A |
| chr9_-_128067310 | 0.66 |
ENST00000373078.5
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr5_+_76850001 | 0.65 |
ENST00000513010.5
ENST00000317593.9 |
S100Z
|
S100 calcium binding protein Z |
| chr2_-_164841208 | 0.64 |
ENST00000439313.5
|
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr1_-_19799872 | 0.63 |
ENST00000294543.11
|
TMCO4
|
transmembrane and coiled-coil domains 4 |
| chr1_-_153977623 | 0.63 |
ENST00000356648.5
|
JTB
|
jumping translocation breakpoint |
| chr2_-_178804623 | 0.63 |
ENST00000359218.10
ENST00000342175.11 |
TTN
|
titin |
| chr2_-_74472643 | 0.63 |
ENST00000258105.8
ENST00000409710.1 |
MRPL53
|
mitochondrial ribosomal protein L53 |
| chr1_+_10399054 | 0.62 |
ENST00000270776.13
ENST00000483936.5 |
PGD
|
phosphogluconate dehydrogenase |
| chr3_+_39329700 | 0.62 |
ENST00000326306.5
|
CCR8
|
C-C motif chemokine receptor 8 |
| chr8_-_92103270 | 0.62 |
ENST00000518832.1
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr12_-_55688891 | 0.61 |
ENST00000557555.2
|
ITGA7
|
integrin subunit alpha 7 |
| chr11_-_111379268 | 0.61 |
ENST00000393067.8
|
POU2AF1
|
POU class 2 homeobox associating factor 1 |
| chr1_+_209827964 | 0.61 |
ENST00000491415.7
|
UTP25
|
UTP25 small subunit processor component |
| chr3_+_39329764 | 0.61 |
ENST00000414803.1
|
CCR8
|
C-C motif chemokine receptor 8 |
| chr7_+_100539188 | 0.59 |
ENST00000300176.9
|
AGFG2
|
ArfGAP with FG repeats 2 |
| chr11_+_810227 | 0.58 |
ENST00000530398.1
|
RPLP2
|
ribosomal protein lateral stalk subunit P2 |
| chr11_-_62754141 | 0.58 |
ENST00000527994.1
ENST00000394807.5 ENST00000673933.1 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr20_+_36091803 | 0.57 |
ENST00000432589.5
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr11_+_10305065 | 0.57 |
ENST00000534464.1
ENST00000278175.10 ENST00000530439.1 ENST00000524948.5 ENST00000528655.5 ENST00000526492.4 ENST00000525063.2 |
ADM
|
adrenomedullin |
| chr12_-_56300358 | 0.57 |
ENST00000550655.5
ENST00000548567.5 ENST00000551430.6 |
CS
|
citrate synthase |
| chr12_-_213338 | 0.55 |
ENST00000424061.6
|
SLC6A12
|
solute carrier family 6 member 12 |
| chr11_+_87037820 | 0.54 |
ENST00000340353.11
|
TMEM135
|
transmembrane protein 135 |
| chr9_+_92974476 | 0.54 |
ENST00000337352.10
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr11_+_67403887 | 0.52 |
ENST00000526387.5
|
TBC1D10C
|
TBC1 domain family member 10C |
| chr2_+_32165841 | 0.52 |
ENST00000357055.7
ENST00000435660.5 ENST00000440718.5 ENST00000379343.6 ENST00000282587.9 ENST00000406369.2 |
SLC30A6
|
solute carrier family 30 member 6 |
| chr11_+_844406 | 0.51 |
ENST00000397404.5
|
TSPAN4
|
tetraspanin 4 |
| chr18_-_69956924 | 0.50 |
ENST00000581982.5
ENST00000280200.8 |
CD226
|
CD226 molecule |
| chr5_+_80488094 | 0.50 |
ENST00000282226.5
|
FAM151B
|
family with sequence similarity 151 member B |
| chr16_+_4624811 | 0.49 |
ENST00000415496.5
ENST00000262370.12 ENST00000587747.5 ENST00000399577.9 ENST00000588994.5 ENST00000586183.5 |
MGRN1
|
mahogunin ring finger 1 |
| chr19_+_1077394 | 0.49 |
ENST00000590577.2
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr17_+_28720552 | 0.49 |
ENST00000472628.1
ENST00000578181.1 |
RPL23A
|
ribosomal protein L23a |
| chr21_-_42010327 | 0.48 |
ENST00000398505.7
ENST00000449949.5 ENST00000310826.10 ENST00000398499.5 ENST00000398497.2 ENST00000398511.3 |
ZBTB21
|
zinc finger and BTB domain containing 21 |
| chr1_-_153977260 | 0.48 |
ENST00000428469.1
|
JTB
|
jumping translocation breakpoint |
| chr11_-_82900406 | 0.48 |
ENST00000313010.8
ENST00000393399.6 ENST00000680437.1 |
PRCP
|
prolylcarboxypeptidase |
| chr15_+_88467692 | 0.48 |
ENST00000325844.9
ENST00000353598.6 |
MRPS11
|
mitochondrial ribosomal protein S11 |
| chr2_+_206765578 | 0.48 |
ENST00000403094.3
ENST00000402774.8 |
FASTKD2
|
FAST kinase domains 2 |
| chr19_+_15737985 | 0.47 |
ENST00000641646.1
|
OR10H3
|
olfactory receptor family 10 subfamily H member 3 |
| chr1_+_179294173 | 0.47 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chr12_-_6568256 | 0.47 |
ENST00000382421.7
ENST00000545200.5 ENST00000399466.6 ENST00000322166.10 ENST00000536124.5 ENST00000540228.1 ENST00000542867.5 ENST00000545492.5 ENST00000545915.5 ENST00000620535.4 ENST00000617555.4 |
NOP2
|
NOP2 nucleolar protein |
| chr5_-_181238195 | 0.47 |
ENST00000509148.1
|
RACK1
|
receptor for activated C kinase 1 |
| chr12_-_213656 | 0.46 |
ENST00000359674.8
|
SLC6A12
|
solute carrier family 6 member 12 |
| chr12_-_214146 | 0.46 |
ENST00000684302.1
|
SLC6A12
|
solute carrier family 6 member 12 |
| chr17_+_8310220 | 0.45 |
ENST00000583529.1
ENST00000361926.8 |
ARHGEF15
|
Rho guanine nucleotide exchange factor 15 |
| chr2_+_27032938 | 0.45 |
ENST00000238788.14
ENST00000404032.7 |
TMEM214
|
transmembrane protein 214 |
| chr17_+_79022908 | 0.44 |
ENST00000354124.7
ENST00000580454.5 |
C1QTNF1
|
C1q and TNF related 1 |
| chr14_-_80315876 | 0.44 |
ENST00000553594.1
|
DIO2
|
iodothyronine deiodinase 2 |
| chr14_-_94504222 | 0.44 |
ENST00000556881.5
|
SERPINA12
|
serpin family A member 12 |
| chr16_-_31094727 | 0.43 |
ENST00000300851.10
ENST00000394975.3 |
VKORC1
|
vitamin K epoxide reductase complex subunit 1 |
| chr7_+_2647703 | 0.43 |
ENST00000403167.5
|
TTYH3
|
tweety family member 3 |
| chr12_+_51888217 | 0.43 |
ENST00000340970.8
|
ANKRD33
|
ankyrin repeat domain 33 |
| chr11_+_119168360 | 0.43 |
ENST00000422249.5
|
NLRX1
|
NLR family member X1 |
| chr19_-_54339146 | 0.43 |
ENST00000291759.5
|
LILRA4
|
leukocyte immunoglobulin like receptor A4 |
| chr6_-_30672984 | 0.42 |
ENST00000415603.1
ENST00000376442.8 |
DHX16
|
DEAH-box helicase 16 |
| chr10_+_99732211 | 0.42 |
ENST00000370476.10
ENST00000370472.4 |
CUTC
|
cutC copper transporter |
| chr19_-_45370384 | 0.42 |
ENST00000485403.6
ENST00000586856.1 ENST00000586131.6 ENST00000391945.10 ENST00000684407.1 ENST00000391944.8 |
ERCC2
|
ERCC excision repair 2, TFIIH core complex helicase subunit |
| chr22_+_35717872 | 0.41 |
ENST00000249044.2
|
APOL5
|
apolipoprotein L5 |
| chr21_-_44920892 | 0.41 |
ENST00000397846.7
ENST00000652462.1 ENST00000302347.10 ENST00000524251.1 |
ITGB2
|
integrin subunit beta 2 |
| chr1_+_26177482 | 0.41 |
ENST00000361530.11
ENST00000374253.9 |
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr7_-_75073774 | 0.41 |
ENST00000610322.5
|
RCC1L
|
RCC1 like |
| chr19_-_48617797 | 0.40 |
ENST00000546623.5
ENST00000084795.9 |
RPL18
|
ribosomal protein L18 |
| chr11_+_1850896 | 0.40 |
ENST00000676039.1
|
LSP1
|
lymphocyte specific protein 1 |
| chr17_-_81637049 | 0.40 |
ENST00000374747.9
ENST00000331134.11 |
NPLOC4
|
NPL4 homolog, ubiquitin recognition factor |
| chr19_-_17255448 | 0.40 |
ENST00000594059.1
|
ENSG00000269095.1
|
novel protein |
| chr8_+_38207586 | 0.40 |
ENST00000521311.1
|
BAG4
|
BAG cochaperone 4 |
| chr3_-_127590707 | 0.40 |
ENST00000296210.11
ENST00000355552.8 |
TPRA1
|
transmembrane protein adipocyte associated 1 |
| chr2_+_216081866 | 0.39 |
ENST00000433112.5
ENST00000454545.5 ENST00000295658.8 ENST00000455479.5 ENST00000406027.2 ENST00000437356.7 |
TMEM169
|
transmembrane protein 169 |
| chr19_-_57916603 | 0.39 |
ENST00000599251.1
ENST00000598526.5 |
ZNF417
|
zinc finger protein 417 |
| chr14_+_20223710 | 0.38 |
ENST00000315519.3
|
OR11H6
|
olfactory receptor family 11 subfamily H member 6 |
| chr16_+_21612071 | 0.38 |
ENST00000562961.1
|
METTL9
|
methyltransferase like 9 |
| chr1_+_152786214 | 0.38 |
ENST00000368771.1
ENST00000368770.4 |
LCE1E
|
late cornified envelope 1E |
| chrX_+_108091752 | 0.38 |
ENST00000457035.5
ENST00000372232.8 |
ATG4A
|
autophagy related 4A cysteine peptidase |
| chr4_-_39366342 | 0.38 |
ENST00000503784.1
ENST00000349703.7 ENST00000381897.5 |
RFC1
|
replication factor C subunit 1 |
| chr1_+_23791203 | 0.38 |
ENST00000374503.7
ENST00000374502.7 |
LYPLA2
|
lysophospholipase 2 |
| chr4_+_102869332 | 0.37 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr11_-_6027741 | 0.36 |
ENST00000641938.1
|
OR56A1
|
olfactory receptor family 56 subfamily A member 1 |
| chr3_+_101561833 | 0.36 |
ENST00000309922.7
|
TRMT10C
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr2_-_218282098 | 0.36 |
ENST00000425694.5
|
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr12_+_51888083 | 0.36 |
ENST00000301190.11
|
ANKRD33
|
ankyrin repeat domain 33 |
| chr3_-_114454683 | 0.36 |
ENST00000470311.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr19_+_15673069 | 0.36 |
ENST00000550308.6
ENST00000551607.5 |
CYP4F12
|
cytochrome P450 family 4 subfamily F member 12 |
| chr21_-_44920918 | 0.35 |
ENST00000522688.5
|
ITGB2
|
integrin subunit beta 2 |
| chr17_-_67996428 | 0.35 |
ENST00000580729.3
|
C17orf58
|
chromosome 17 open reading frame 58 |
| chr12_-_214082 | 0.35 |
ENST00000535347.5
ENST00000536824.5 |
SLC6A12
|
solute carrier family 6 member 12 |
| chr2_+_167187364 | 0.35 |
ENST00000672671.1
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr3_+_101561926 | 0.35 |
ENST00000495642.1
|
TRMT10C
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr19_+_8418386 | 0.35 |
ENST00000602117.1
|
MARCHF2
|
membrane associated ring-CH-type finger 2 |
| chr12_+_123458092 | 0.35 |
ENST00000526639.3
|
SNRNP35
|
small nuclear ribonucleoprotein U11/U12 subunit 35 |
| chr11_+_62885177 | 0.34 |
ENST00000539507.1
|
SLC3A2
|
solute carrier family 3 member 2 |
| chr4_+_38867789 | 0.34 |
ENST00000358869.5
|
FAM114A1
|
family with sequence similarity 114 member A1 |
| chr5_+_172983763 | 0.34 |
ENST00000519374.6
ENST00000519911.5 |
ATP6V0E1
|
ATPase H+ transporting V0 subunit e1 |
| chr1_+_35869750 | 0.34 |
ENST00000373206.5
|
AGO1
|
argonaute RISC component 1 |
| chr14_-_73458519 | 0.34 |
ENST00000356296.8
ENST00000557597.5 ENST00000554394.5 ENST00000555238.6 ENST00000555987.5 ENST00000555394.5 ENST00000554546.5 |
NUMB
|
NUMB endocytic adaptor protein |
| chr7_+_16661182 | 0.33 |
ENST00000446596.5
ENST00000452975.6 ENST00000438834.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr8_-_28083850 | 0.33 |
ENST00000418860.1
|
NUGGC
|
nuclear GTPase, germinal center associated |
| chr21_+_42199686 | 0.33 |
ENST00000398457.6
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr19_+_57849834 | 0.32 |
ENST00000339656.8
ENST00000423137.1 |
ZNF587
|
zinc finger protein 587 |
| chr3_+_46880242 | 0.32 |
ENST00000418619.5
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr1_+_23791138 | 0.32 |
ENST00000374514.8
ENST00000420982.5 ENST00000374505.6 |
LYPLA2
|
lysophospholipase 2 |
| chr12_+_53241884 | 0.32 |
ENST00000635789.1
|
ENSG00000283536.2
|
novel protein |
| chr22_-_33572227 | 0.32 |
ENST00000674780.1
|
LARGE1
|
LARGE xylosyl- and glucuronyltransferase 1 |
| chr14_-_59484317 | 0.31 |
ENST00000247194.9
|
L3HYPDH
|
trans-L-3-hydroxyproline dehydratase |
| chr19_+_52274443 | 0.31 |
ENST00000600821.5
ENST00000595149.5 ENST00000595000.5 ENST00000593612.1 |
ZNF766
|
zinc finger protein 766 |
| chr1_+_156369202 | 0.31 |
ENST00000537040.6
|
RHBG
|
Rh family B glycoprotein |
| chr14_-_70359630 | 0.30 |
ENST00000389912.7
|
COX16
|
cytochrome c oxidase assembly factor COX16 |
| chrX_-_72714181 | 0.30 |
ENST00000339490.7
ENST00000541944.5 ENST00000373539.3 |
PHKA1
|
phosphorylase kinase regulatory subunit alpha 1 |
| chr11_+_827248 | 0.30 |
ENST00000527089.5
ENST00000530183.1 |
CRACR2B
|
calcium release activated channel regulator 2B |
| chr17_+_78187317 | 0.30 |
ENST00000409257.9
ENST00000591256.5 ENST00000589256.5 ENST00000588800.5 ENST00000591952.5 ENST00000327898.9 ENST00000586542.5 ENST00000586731.1 |
AFMID
|
arylformamidase |
| chr16_+_71626149 | 0.29 |
ENST00000567566.1
|
MARVELD3
|
MARVEL domain containing 3 |
| chr9_-_113397356 | 0.29 |
ENST00000452726.1
|
ALAD
|
aminolevulinate dehydratase |
| chrX_+_108091665 | 0.29 |
ENST00000345734.7
|
ATG4A
|
autophagy related 4A cysteine peptidase |
| chr16_+_53099100 | 0.29 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr1_+_223701607 | 0.29 |
ENST00000434648.5
|
CAPN2
|
calpain 2 |
| chr7_+_45027634 | 0.28 |
ENST00000381112.7
ENST00000474617.1 |
CCM2
|
CCM2 scaffold protein |
| chr1_+_13303539 | 0.28 |
ENST00000437300.2
|
PRAMEF33
|
PRAME family member 33 |
| chr9_-_41189310 | 0.28 |
ENST00000456520.5
ENST00000377391.8 ENST00000613716.4 ENST00000617933.1 |
CBWD6
|
COBW domain containing 6 |
| chr12_+_6970904 | 0.28 |
ENST00000599672.6
ENST00000539196.2 |
EMG1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr9_+_68241854 | 0.28 |
ENST00000616550.4
ENST00000618217.4 ENST00000377342.9 ENST00000478048.5 ENST00000360171.11 |
CBWD3
|
COBW domain containing 3 |
| chr19_+_54105923 | 0.28 |
ENST00000420296.1
|
NDUFA3
|
NADH:ubiquinone oxidoreductase subunit A3 |
| chr1_-_153946652 | 0.27 |
ENST00000361217.9
|
DENND4B
|
DENN domain containing 4B |
| chr21_-_44920855 | 0.27 |
ENST00000397854.7
|
ITGB2
|
integrin subunit beta 2 |
| chr20_+_19758245 | 0.27 |
ENST00000255006.12
|
RIN2
|
Ras and Rab interactor 2 |
| chr7_-_138679045 | 0.27 |
ENST00000419765.4
|
SVOPL
|
SVOP like |
| chr2_+_167187283 | 0.26 |
ENST00000409605.1
ENST00000409273.6 |
XIRP2
|
xin actin binding repeat containing 2 |
| chr16_+_28863023 | 0.26 |
ENST00000566209.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr7_-_73738831 | 0.26 |
ENST00000395147.9
ENST00000437775.7 |
ABHD11
|
abhydrolase domain containing 11 |
| chr1_-_12945416 | 0.26 |
ENST00000415464.6
|
PRAMEF6
|
PRAME family member 6 |
| chr14_-_73458248 | 0.26 |
ENST00000555307.5
ENST00000554818.5 |
NUMB
|
NUMB endocytic adaptor protein |
| chr4_+_38867677 | 0.26 |
ENST00000510213.5
ENST00000515037.5 |
FAM114A1
|
family with sequence similarity 114 member A1 |
| chr17_-_78187036 | 0.26 |
ENST00000590862.5
ENST00000590430.5 ENST00000301634.12 ENST00000586613.1 |
TK1
|
thymidine kinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1H2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.1 | GO:1901569 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.5 | 2.1 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.4 | 2.7 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.4 | 1.3 | GO:1903576 | 'de novo' UMP biosynthetic process(GO:0044205) response to L-arginine(GO:1903576) |
| 0.4 | 1.6 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.4 | 1.1 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.3 | 1.6 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.3 | 3.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 6.9 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.2 | 0.7 | GO:0000964 | mitochondrial RNA 5'-end processing(GO:0000964) |
| 0.2 | 1.6 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.2 | 0.7 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 | 0.7 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.2 | 0.7 | GO:0044115 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.2 | 1.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.6 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 2.5 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 1.2 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 0.7 | GO:0032796 | uropod organization(GO:0032796) |
| 0.1 | 0.4 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 1.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 1.8 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.1 | 0.5 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 1.0 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.1 | 0.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.9 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.3 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.1 | 0.7 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.5 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.2 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.1 | 0.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.8 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 0.7 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 0.8 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.7 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.9 | GO:2000332 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.7 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.5 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 1.8 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 0.7 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.4 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.2 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.4 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 | 0.7 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.5 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.5 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.1 | 0.6 | GO:0097646 | progesterone biosynthetic process(GO:0006701) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.7 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.0 | 0.5 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 2.2 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.3 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 1.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.6 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.2 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.9 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.7 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.5 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.8 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.8 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.3 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 1.0 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.8 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.8 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 0.3 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 2.9 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 1.0 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.3 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.3 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 2.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 1.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0071757 | hexameric IgM immunoglobulin complex(GO:0071757) |
| 0.4 | 1.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 0.7 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.2 | 0.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.7 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 1.0 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.4 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.1 | 0.7 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.2 | GO:0070701 | mucus layer(GO:0070701) |
| 0.0 | 0.5 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 1.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 3.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 4.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 1.0 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 1.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.7 | 2.1 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.5 | 1.6 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.5 | 2.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.5 | 3.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.4 | 1.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.4 | 2.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.3 | 1.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.3 | 1.3 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.2 | 0.7 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.2 | 0.4 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.2 | 0.6 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.2 | 0.8 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.2 | 1.5 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.2 | 1.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 1.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 0.5 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 0.9 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.7 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.6 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.4 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.1 | 0.5 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.3 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.1 | 1.0 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 0.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 1.0 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.8 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 1.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 1.0 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 1.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.2 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.1 | 0.4 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 1.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.4 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 1.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.4 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.7 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.3 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.0 | 0.2 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 1.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 1.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 2.3 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 3.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.5 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 2.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |