Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
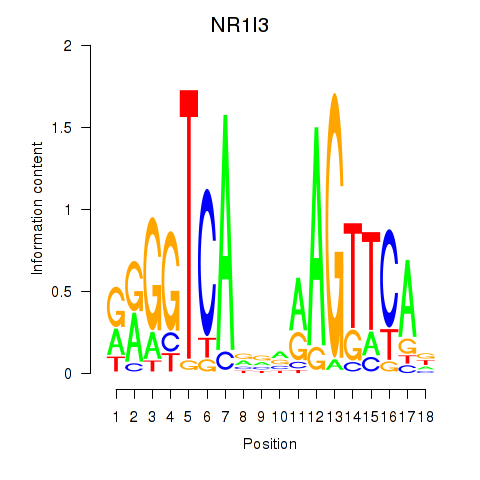
Results for NR1I3
Z-value: 0.37
Transcription factors associated with NR1I3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I3
|
ENSG00000143257.12 | nuclear receptor subfamily 1 group I member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I3 | hg38_v1_chr1_-_161238196_161238210 | 0.19 | 3.0e-01 | Click! |
Activity profile of NR1I3 motif
Sorted Z-values of NR1I3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_154405326 | 0.80 |
ENST00000368485.8
|
IL6R
|
interleukin 6 receptor |
| chr1_+_154405193 | 0.76 |
ENST00000622330.4
ENST00000344086.8 |
IL6R
|
interleukin 6 receptor |
| chr1_-_205321737 | 0.70 |
ENST00000367157.6
|
NUAK2
|
NUAK family kinase 2 |
| chr11_-_61356793 | 0.61 |
ENST00000539890.1
|
CYB561A3
|
cytochrome b561 family member A3 |
| chr1_+_153357846 | 0.59 |
ENST00000368738.4
|
S100A9
|
S100 calcium binding protein A9 |
| chr1_+_154405573 | 0.59 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr7_-_56092932 | 0.58 |
ENST00000446428.5
ENST00000432123.5 ENST00000297373.7 |
PHKG1
|
phosphorylase kinase catalytic subunit gamma 1 |
| chr8_+_11494367 | 0.55 |
ENST00000259089.9
ENST00000529894.1 |
BLK
|
BLK proto-oncogene, Src family tyrosine kinase |
| chr8_+_32614361 | 0.54 |
ENST00000522569.1
|
NRG1
|
neuregulin 1 |
| chr7_-_56092974 | 0.50 |
ENST00000452681.6
ENST00000537360.5 |
PHKG1
|
phosphorylase kinase catalytic subunit gamma 1 |
| chr14_-_102509713 | 0.45 |
ENST00000286918.9
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr11_+_62789124 | 0.43 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr6_+_96521796 | 0.42 |
ENST00000369278.5
|
UFL1
|
UFM1 specific ligase 1 |
| chrX_-_33211312 | 0.40 |
ENST00000420596.5
ENST00000448370.5 |
DMD
|
dystrophin |
| chr14_+_24130659 | 0.39 |
ENST00000267426.6
|
FITM1
|
fat storage inducing transmembrane protein 1 |
| chr11_-_72041945 | 0.38 |
ENST00000543009.5
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr11_-_64996963 | 0.37 |
ENST00000301887.9
ENST00000534177.1 |
BATF2
|
basic leucine zipper ATF-like transcription factor 2 |
| chr6_+_32439866 | 0.37 |
ENST00000374982.5
ENST00000395388.7 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr5_-_177510348 | 0.37 |
ENST00000377112.8
ENST00000501403.6 ENST00000312943.10 |
DOK3
|
docking protein 3 |
| chr12_+_53380639 | 0.36 |
ENST00000426431.2
|
SP1
|
Sp1 transcription factor |
| chr5_+_139675471 | 0.36 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr14_-_102509798 | 0.36 |
ENST00000560748.5
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr8_-_142752510 | 0.35 |
ENST00000359228.4
|
LYPD2
|
LY6/PLAUR domain containing 2 |
| chr7_-_73738831 | 0.35 |
ENST00000395147.9
ENST00000437775.7 |
ABHD11
|
abhydrolase domain containing 11 |
| chr7_-_73738792 | 0.34 |
ENST00000222800.8
ENST00000458339.6 |
ABHD11
|
abhydrolase domain containing 11 |
| chr16_+_67873036 | 0.32 |
ENST00000358933.10
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr18_-_23586422 | 0.32 |
ENST00000269228.10
|
NPC1
|
NPC intracellular cholesterol transporter 1 |
| chr15_+_90266317 | 0.32 |
ENST00000622269.1
ENST00000411845.3 |
ENSG00000275674.1
NGRN
|
novel protein neugrin, neurite outgrowth associated |
| chr1_+_56854764 | 0.31 |
ENST00000361249.4
|
C8A
|
complement C8 alpha chain |
| chr19_-_50833187 | 0.31 |
ENST00000598673.1
|
KLK15
|
kallikrein related peptidase 15 |
| chr1_-_145918485 | 0.31 |
ENST00000537888.1
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr14_+_22281097 | 0.31 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr5_-_39270623 | 0.31 |
ENST00000512138.1
ENST00000646045.2 |
FYB1
|
FYN binding protein 1 |
| chr5_-_177509814 | 0.31 |
ENST00000510898.7
ENST00000502885.5 ENST00000506493.5 |
DOK3
|
docking protein 3 |
| chr10_+_133160401 | 0.31 |
ENST00000684739.1
ENST00000682765.1 ENST00000683144.1 ENST00000683259.1 ENST00000682399.1 ENST00000368571.3 |
KNDC1
|
kinase non-catalytic C-lobe domain containing 1 |
| chr11_-_72070050 | 0.29 |
ENST00000535087.5
ENST00000535838.5 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr3_-_108757407 | 0.29 |
ENST00000295755.7
|
RETNLB
|
resistin like beta |
| chr11_+_113908983 | 0.29 |
ENST00000537778.5
|
HTR3B
|
5-hydroxytryptamine receptor 3B |
| chr1_-_145918685 | 0.28 |
ENST00000369306.8
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr17_-_29294141 | 0.28 |
ENST00000225388.9
|
NUFIP2
|
nuclear FMR1 interacting protein 2 |
| chr10_-_80173014 | 0.27 |
ENST00000437799.1
|
ANXA11
|
annexin A11 |
| chr14_+_21057822 | 0.27 |
ENST00000308227.2
|
RNASE8
|
ribonuclease A family member 8 |
| chr6_+_108166015 | 0.26 |
ENST00000368986.9
|
NR2E1
|
nuclear receptor subfamily 2 group E member 1 |
| chr7_-_99784175 | 0.26 |
ENST00000651514.1
ENST00000336411.7 ENST00000415003.1 ENST00000354593.6 |
CYP3A4
|
cytochrome P450 family 3 subfamily A member 4 |
| chr16_-_66934144 | 0.26 |
ENST00000568572.5
|
CIAO2B
|
cytosolic iron-sulfur assembly component 2B |
| chrX_-_111412162 | 0.25 |
ENST00000637570.1
ENST00000356220.8 ENST00000636035.2 ENST00000637453.1 ENST00000635795.1 |
DCX
|
doublecortin |
| chr3_-_71753582 | 0.25 |
ENST00000295612.7
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr17_-_35063648 | 0.25 |
ENST00000394597.7
|
RFFL
|
ring finger and FYVE like domain containing E3 ubiquitin protein ligase |
| chr1_-_45542726 | 0.24 |
ENST00000676549.1
|
PRDX1
|
peroxiredoxin 1 |
| chr17_-_29294107 | 0.24 |
ENST00000579665.1
|
NUFIP2
|
nuclear FMR1 interacting protein 2 |
| chr19_+_58408735 | 0.23 |
ENST00000599238.5
ENST00000322834.7 |
ZNF584
|
zinc finger protein 584 |
| chr7_-_99735093 | 0.23 |
ENST00000611620.4
ENST00000620220.6 ENST00000336374.4 |
CYP3A7-CYP3A51P
CYP3A7
|
CYP3A7-CYP3A51P readthrough cytochrome P450 family 3 subfamily A member 7 |
| chr4_+_70028452 | 0.23 |
ENST00000530128.5
ENST00000381057.3 ENST00000673563.1 |
HTN3
|
histatin 3 |
| chr2_-_68952880 | 0.23 |
ENST00000481498.1
ENST00000328895.9 |
GKN2
|
gastrokine 2 |
| chr16_-_66934362 | 0.22 |
ENST00000567511.1
ENST00000422424.7 |
CIAO2B
|
cytosolic iron-sulfur assembly component 2B |
| chr22_-_23974441 | 0.22 |
ENST00000398344.9
ENST00000403754.7 ENST00000430101.2 |
DDT
|
D-dopachrome tautomerase |
| chr7_-_99784248 | 0.22 |
ENST00000652018.1
|
CYP3A4
|
cytochrome P450 family 3 subfamily A member 4 |
| chr7_+_143381561 | 0.22 |
ENST00000354434.8
|
ZYX
|
zyxin |
| chr11_-_83682260 | 0.22 |
ENST00000398304.5
ENST00000420775.6 |
DLG2
|
discs large MAGUK scaffold protein 2 |
| chr1_+_40817190 | 0.22 |
ENST00000443478.3
|
KCNQ4
|
potassium voltage-gated channel subfamily Q member 4 |
| chr12_-_118372883 | 0.22 |
ENST00000542532.5
ENST00000392533.8 |
TAOK3
|
TAO kinase 3 |
| chr12_-_53335737 | 0.21 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr11_+_63369779 | 0.21 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chr16_+_67246895 | 0.21 |
ENST00000566345.6
|
SLC9A5
|
solute carrier family 9 member A5 |
| chr2_-_73269483 | 0.20 |
ENST00000295133.9
|
FBXO41
|
F-box protein 41 |
| chr12_-_75207998 | 0.20 |
ENST00000550433.5
ENST00000548513.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr20_+_36461303 | 0.19 |
ENST00000475894.5
|
DLGAP4
|
DLG associated protein 4 |
| chr19_+_58408633 | 0.19 |
ENST00000306910.9
ENST00000598901.1 ENST00000593920.5 ENST00000596281.1 |
ZNF584
|
zinc finger protein 584 |
| chr18_-_21703688 | 0.19 |
ENST00000584464.1
ENST00000578270.5 |
ABHD3
|
abhydrolase domain containing 3, phospholipase |
| chr8_-_108787563 | 0.19 |
ENST00000297459.4
|
TMEM74
|
transmembrane protein 74 |
| chr19_+_51478533 | 0.19 |
ENST00000396477.4
|
CEACAM18
|
CEA cell adhesion molecule 18 |
| chr5_+_139675436 | 0.19 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr2_+_70087468 | 0.18 |
ENST00000303577.7
|
PCBP1
|
poly(rC) binding protein 1 |
| chr9_+_34179005 | 0.18 |
ENST00000625521.2
ENST00000379186.8 ENST00000297661.9 ENST00000626262.2 |
UBAP1
|
ubiquitin associated protein 1 |
| chr7_+_143381907 | 0.18 |
ENST00000392910.6
|
ZYX
|
zyxin |
| chr11_-_89065969 | 0.18 |
ENST00000305447.5
|
GRM5
|
glutamate metabotropic receptor 5 |
| chr9_+_5450503 | 0.18 |
ENST00000381573.8
ENST00000381577.4 |
CD274
|
CD274 molecule |
| chr6_+_70566892 | 0.18 |
ENST00000370474.4
|
SDHAF4
|
succinate dehydrogenase complex assembly factor 4 |
| chrX_+_48786578 | 0.17 |
ENST00000376670.9
|
GATA1
|
GATA binding protein 1 |
| chr3_+_52316319 | 0.17 |
ENST00000420323.7
|
DNAH1
|
dynein axonemal heavy chain 1 |
| chr1_+_152947154 | 0.17 |
ENST00000636302.1
|
SPRR5
|
small proline rich protein 5 |
| chr11_-_47848539 | 0.17 |
ENST00000526870.1
|
NUP160
|
nucleoporin 160 |
| chr10_-_80172839 | 0.16 |
ENST00000265447.8
|
ANXA11
|
annexin A11 |
| chr5_-_136193143 | 0.16 |
ENST00000607574.2
|
SMIM32
|
small integral membrane protein 32 |
| chr15_-_42051190 | 0.16 |
ENST00000399518.3
|
PLA2G4E
|
phospholipase A2 group IVE |
| chr1_+_160127672 | 0.15 |
ENST00000447527.1
|
ATP1A2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr16_+_2988256 | 0.15 |
ENST00000573315.2
|
LINC00514
|
long intergenic non-protein coding RNA 514 |
| chr1_-_151146643 | 0.15 |
ENST00000613223.1
|
SEMA6C
|
semaphorin 6C |
| chr14_-_77320813 | 0.15 |
ENST00000682467.1
|
POMT2
|
protein O-mannosyltransferase 2 |
| chr3_-_56468346 | 0.14 |
ENST00000288221.11
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr10_+_102503956 | 0.13 |
ENST00000369902.8
|
SUFU
|
SUFU negative regulator of hedgehog signaling |
| chr16_+_527698 | 0.13 |
ENST00000219611.7
ENST00000562370.5 ENST00000568988.5 |
CAPN15
|
calpain 15 |
| chr1_+_43935807 | 0.13 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr1_+_81699665 | 0.13 |
ENST00000359929.7
|
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr12_-_108598002 | 0.12 |
ENST00000549447.1
|
TMEM119
|
transmembrane protein 119 |
| chr1_-_225428813 | 0.12 |
ENST00000338179.6
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr14_-_77320741 | 0.12 |
ENST00000682795.1
ENST00000682247.1 |
POMT2
|
protein O-mannosyltransferase 2 |
| chr19_-_48390847 | 0.12 |
ENST00000597017.5
|
KDELR1
|
KDEL endoplasmic reticulum protein retention receptor 1 |
| chr1_+_32465046 | 0.12 |
ENST00000609129.2
|
ZBTB8B
|
zinc finger and BTB domain containing 8B |
| chr11_-_7963646 | 0.12 |
ENST00000328600.3
|
NLRP10
|
NLR family pyrin domain containing 10 |
| chr11_+_933553 | 0.11 |
ENST00000534485.5
|
AP2A2
|
adaptor related protein complex 2 subunit alpha 2 |
| chr14_+_52707192 | 0.11 |
ENST00000445930.7
ENST00000555339.5 ENST00000556813.1 |
PSMC6
|
proteasome 26S subunit, ATPase 6 |
| chr14_-_21537064 | 0.11 |
ENST00000541965.1
|
SALL2
|
spalt like transcription factor 2 |
| chr11_-_61357239 | 0.11 |
ENST00000536915.5
|
CYB561A3
|
cytochrome b561 family member A3 |
| chr1_+_27392612 | 0.11 |
ENST00000374024.4
|
GPR3
|
G protein-coupled receptor 3 |
| chr7_-_143882808 | 0.11 |
ENST00000460532.5
ENST00000491908.1 |
TCAF1
|
TRPM8 channel associated factor 1 |
| chr14_-_21536884 | 0.11 |
ENST00000546363.5
|
SALL2
|
spalt like transcription factor 2 |
| chr6_+_33454543 | 0.10 |
ENST00000621915.1
ENST00000395064.3 |
ZBTB9
|
zinc finger and BTB domain containing 9 |
| chr10_+_86654581 | 0.10 |
ENST00000443292.1
|
OPN4
|
opsin 4 |
| chr11_-_57322300 | 0.10 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr6_-_31541937 | 0.10 |
ENST00000456662.5
ENST00000431908.5 ENST00000456976.5 ENST00000428450.5 ENST00000418897.5 ENST00000396172.6 ENST00000419020.1 ENST00000428098.5 |
DDX39B
|
DExD-box helicase 39B |
| chr19_-_6604083 | 0.10 |
ENST00000597430.2
|
CD70
|
CD70 molecule |
| chr12_-_21403637 | 0.10 |
ENST00000450590.5
ENST00000453443.5 |
SLCO1A2
|
solute carrier organic anion transporter family member 1A2 |
| chr8_-_52714414 | 0.09 |
ENST00000435644.6
ENST00000518710.5 ENST00000025008.10 ENST00000517963.1 |
RB1CC1
|
RB1 inducible coiled-coil 1 |
| chr1_-_9026386 | 0.09 |
ENST00000400906.2
|
SLC2A7
|
solute carrier family 2 member 7 |
| chr14_-_77320855 | 0.09 |
ENST00000556394.2
ENST00000261534.9 |
POMT2
|
protein O-mannosyltransferase 2 |
| chr13_-_94479671 | 0.09 |
ENST00000377028.10
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr11_-_47848313 | 0.09 |
ENST00000530326.5
ENST00000528071.5 |
NUP160
|
nucleoporin 160 |
| chr11_-_89065945 | 0.09 |
ENST00000393294.3
|
GRM5
|
glutamate metabotropic receptor 5 |
| chr16_-_74985070 | 0.09 |
ENST00000616369.4
ENST00000262144.11 |
WDR59
|
WD repeat domain 59 |
| chr2_-_39120383 | 0.08 |
ENST00000395038.6
|
SOS1
|
SOS Ras/Rac guanine nucleotide exchange factor 1 |
| chr1_-_21298988 | 0.08 |
ENST00000481130.6
|
ECE1
|
endothelin converting enzyme 1 |
| chr1_-_155187272 | 0.08 |
ENST00000473363.3
|
ENSG00000273088.2
|
novel protein |
| chr6_-_34146080 | 0.08 |
ENST00000538487.7
ENST00000374181.8 |
GRM4
|
glutamate metabotropic receptor 4 |
| chr19_+_54200849 | 0.08 |
ENST00000626547.2
ENST00000302907.9 ENST00000391752.5 ENST00000402367.5 ENST00000391751.7 |
RPS9
|
ribosomal protein S9 |
| chr17_-_58219227 | 0.07 |
ENST00000581180.2
ENST00000313863.11 ENST00000393119.7 ENST00000678463.1 ENST00000676787.1 ENST00000580127.6 ENST00000585134.2 ENST00000581761.6 |
MKS1
|
MKS transition zone complex subunit 1 |
| chr4_+_37244735 | 0.07 |
ENST00000309447.6
|
NWD2
|
NACHT and WD repeat domain containing 2 |
| chr2_-_86563349 | 0.07 |
ENST00000409727.5
|
CHMP3
|
charged multivesicular body protein 3 |
| chr1_-_158426237 | 0.07 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr11_-_47848467 | 0.07 |
ENST00000378460.6
|
NUP160
|
nucleoporin 160 |
| chr2_-_86563382 | 0.07 |
ENST00000263856.9
|
CHMP3
|
charged multivesicular body protein 3 |
| chr11_-_31812171 | 0.07 |
ENST00000639409.1
ENST00000640975.1 |
PAX6
|
paired box 6 |
| chr17_+_28506233 | 0.07 |
ENST00000577936.1
|
FOXN1
|
forkhead box N1 |
| chr2_-_227164194 | 0.07 |
ENST00000396625.5
|
COL4A4
|
collagen type IV alpha 4 chain |
| chr11_+_197303 | 0.06 |
ENST00000342593.6
|
ODF3
|
outer dense fiber of sperm tails 3 |
| chr11_-_61357304 | 0.06 |
ENST00000542361.5
|
CYB561A3
|
cytochrome b561 family member A3 |
| chr19_-_48637338 | 0.05 |
ENST00000601104.1
ENST00000222122.10 |
DBP
|
D-box binding PAR bZIP transcription factor |
| chr19_+_54201122 | 0.05 |
ENST00000391753.6
ENST00000441429.1 |
RPS9
|
ribosomal protein S9 |
| chr4_-_81214960 | 0.05 |
ENST00000395578.3
ENST00000628926.1 |
PRKG2
|
protein kinase cGMP-dependent 2 |
| chr17_-_40648646 | 0.05 |
ENST00000643318.1
|
SMARCE1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr2_-_68871382 | 0.04 |
ENST00000295379.2
|
BMP10
|
bone morphogenetic protein 10 |
| chr19_-_45792755 | 0.04 |
ENST00000377735.7
ENST00000270223.7 |
DMWD
|
DM1 locus, WD repeat containing |
| chr10_+_18400562 | 0.04 |
ENST00000377315.5
ENST00000650685.1 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr1_+_44808482 | 0.03 |
ENST00000450269.5
ENST00000409335.6 |
BTBD19
|
BTB domain containing 19 |
| chr2_-_86563470 | 0.03 |
ENST00000409225.2
|
CHMP3
|
charged multivesicular body protein 3 |
| chr5_+_53560627 | 0.03 |
ENST00000296684.10
ENST00000506765.1 |
NDUFS4
|
NADH:ubiquinone oxidoreductase subunit S4 |
| chr9_+_37486006 | 0.03 |
ENST00000377792.3
|
POLR1E
|
RNA polymerase I subunit E |
| chr18_-_21704763 | 0.03 |
ENST00000580981.5
ENST00000289119.7 |
ABHD3
|
abhydrolase domain containing 3, phospholipase |
| chrX_+_129738942 | 0.03 |
ENST00000371106.4
|
XPNPEP2
|
X-prolyl aminopeptidase 2 |
| chr14_+_52707178 | 0.03 |
ENST00000612399.4
|
PSMC6
|
proteasome 26S subunit, ATPase 6 |
| chr14_-_21536928 | 0.02 |
ENST00000613414.4
|
SALL2
|
spalt like transcription factor 2 |
| chr16_+_30699155 | 0.02 |
ENST00000262518.9
|
SRCAP
|
Snf2 related CREBBP activator protein |
| chr16_-_46689145 | 0.02 |
ENST00000299138.12
|
VPS35
|
VPS35 retromer complex component |
| chr1_+_43337803 | 0.01 |
ENST00000372470.9
ENST00000413998.7 |
MPL
|
MPL proto-oncogene, thrombopoietin receptor |
| chr12_+_53300027 | 0.01 |
ENST00000549488.5
|
MYG1
|
MYG1 exonuclease |
| chr1_-_151146611 | 0.01 |
ENST00000341697.7
ENST00000368914.8 |
SEMA6C
|
semaphorin 6C |
| chr3_+_122384167 | 0.01 |
ENST00000232125.9
ENST00000477892.5 ENST00000469967.1 |
FAM162A
|
family with sequence similarity 162 member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.4 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.4 | GO:0100057 | regulation of phenotypic switching by transcription from RNA polymerase II promoter(GO:0100057) regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.1 | 0.5 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 0.7 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 0.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.2 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.1 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.1 | 0.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.4 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.4 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.2 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 0.0 | 0.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.2 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.0 | 0.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.5 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.1 | GO:0097536 | thymus epithelium morphogenesis(GO:0097536) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.9 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.1 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.4 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.6 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.4 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.2 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.1 | 0.7 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.5 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.3 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.9 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.2 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 0.2 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 0.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.5 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |