Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
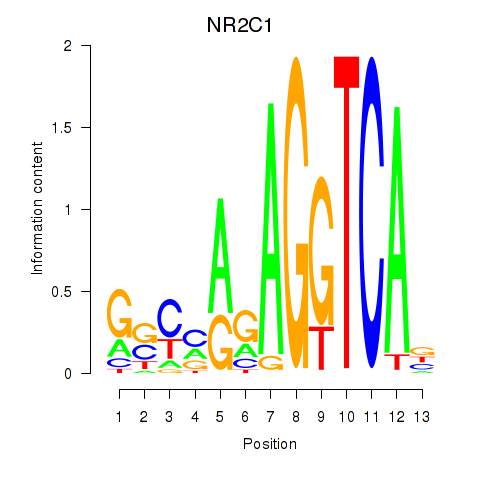
Results for NR2C1
Z-value: 1.30
Transcription factors associated with NR2C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2C1
|
ENSG00000120798.17 | nuclear receptor subfamily 2 group C member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2C1 | hg38_v1_chr12_-_95073580_95073641 | -0.33 | 6.5e-02 | Click! |
Activity profile of NR2C1 motif
Sorted Z-values of NR2C1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_229434086 | 11.97 |
ENST00000366684.7
ENST00000684723.1 ENST00000366683.4 |
ACTA1
|
actin alpha 1, skeletal muscle |
| chr17_+_39665340 | 8.69 |
ENST00000578283.1
ENST00000309889.3 |
TCAP
|
titin-cap |
| chr16_-_4242068 | 6.20 |
ENST00000399609.7
|
SRL
|
sarcalumenin |
| chr1_-_982086 | 5.46 |
ENST00000341290.6
|
PERM1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr17_+_4950147 | 4.83 |
ENST00000522301.5
|
ENO3
|
enolase 3 |
| chr12_+_48106094 | 4.80 |
ENST00000546755.5
ENST00000549366.5 ENST00000642730.1 ENST00000552792.5 |
PFKM
|
phosphofructokinase, muscle |
| chr14_+_104745960 | 4.74 |
ENST00000556623.1
ENST00000555674.1 |
ADSS1
|
adenylosuccinate synthase 1 |
| chr6_-_159727324 | 4.57 |
ENST00000401980.3
ENST00000545162.5 |
SOD2
|
superoxide dismutase 2 |
| chr19_+_16661121 | 4.44 |
ENST00000187762.7
ENST00000599479.1 |
TMEM38A
|
transmembrane protein 38A |
| chr6_-_159726871 | 4.39 |
ENST00000535561.5
|
SOD2
|
superoxide dismutase 2 |
| chr19_-_48363914 | 4.27 |
ENST00000377431.6
ENST00000293261.8 |
TMEM143
|
transmembrane protein 143 |
| chr1_+_160127672 | 3.72 |
ENST00000447527.1
|
ATP1A2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr9_-_35685462 | 3.63 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 |
| chr19_-_48364034 | 3.59 |
ENST00000435956.7
|
TMEM143
|
transmembrane protein 143 |
| chr22_+_31122923 | 3.07 |
ENST00000620191.4
ENST00000412277.6 ENST00000412985.5 ENST00000331075.10 ENST00000420017.5 ENST00000400294.6 ENST00000405300.5 ENST00000404390.7 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr11_+_10450627 | 2.93 |
ENST00000396554.7
ENST00000524866.5 |
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr3_+_37243333 | 2.91 |
ENST00000431105.1
|
GOLGA4
|
golgin A4 |
| chr4_-_110623051 | 2.66 |
ENST00000557119.2
ENST00000644743.1 |
PITX2
|
paired like homeodomain 2 |
| chr15_-_82806054 | 2.63 |
ENST00000541889.1
ENST00000334574.12 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr12_+_109139397 | 2.54 |
ENST00000377854.9
ENST00000377848.7 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr11_+_10450289 | 2.53 |
ENST00000444303.6
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr18_+_7231125 | 2.44 |
ENST00000383467.2
|
LRRC30
|
leucine rich repeat containing 30 |
| chr17_+_14069489 | 2.37 |
ENST00000429152.6
ENST00000261643.8 |
COX10
|
cytochrome c oxidase assembly factor heme A:farnesyltransferase COX10 |
| chr6_+_133240587 | 2.34 |
ENST00000452339.6
|
EYA4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr1_+_43946905 | 2.04 |
ENST00000372343.8
|
IPO13
|
importin 13 |
| chr11_+_112025367 | 2.02 |
ENST00000679614.1
ENST00000679878.1 ENST00000280346.11 ENST00000681339.1 ENST00000681328.1 ENST00000681316.1 ENST00000531306.2 ENST00000680331.1 ENST00000393051.5 |
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr3_+_37243177 | 2.02 |
ENST00000361924.6
ENST00000444882.5 ENST00000356847.8 ENST00000617480.4 ENST00000450863.6 ENST00000429018.5 |
GOLGA4
|
golgin A4 |
| chr3_+_159839847 | 2.00 |
ENST00000445224.6
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr10_+_62049211 | 2.00 |
ENST00000309334.5
|
ARID5B
|
AT-rich interaction domain 5B |
| chr8_-_143953845 | 1.81 |
ENST00000354958.6
|
PLEC
|
plectin |
| chr11_+_64306227 | 1.76 |
ENST00000405666.5
ENST00000468670.2 |
ESRRA
|
estrogen related receptor alpha |
| chr2_+_201451711 | 1.73 |
ENST00000194530.8
ENST00000392249.6 |
STRADB
|
STE20 related adaptor beta |
| chr8_-_144063951 | 1.72 |
ENST00000567871.2
|
OPLAH
|
5-oxoprolinase, ATP-hydrolysing |
| chr10_+_24466487 | 1.70 |
ENST00000396446.5
ENST00000396445.5 ENST00000376451.4 |
KIAA1217
|
KIAA1217 |
| chr3_+_155083889 | 1.69 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase |
| chr11_+_94768331 | 1.66 |
ENST00000317829.12
ENST00000433060.3 |
AMOTL1
|
angiomotin like 1 |
| chr4_+_158672266 | 1.54 |
ENST00000684622.1
ENST00000683483.1 ENST00000684641.1 ENST00000682456.1 ENST00000684627.1 ENST00000511912.6 ENST00000684505.1 ENST00000683305.1 ENST00000684036.1 ENST00000683751.1 ENST00000684129.1 ENST00000307738.5 |
ETFDH
|
electron transfer flavoprotein dehydrogenase |
| chr3_+_155083523 | 1.53 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase |
| chrX_-_112840815 | 1.52 |
ENST00000304758.5
ENST00000371959.9 |
AMOT
|
angiomotin |
| chr15_+_90906741 | 1.45 |
ENST00000557865.5
|
MAN2A2
|
mannosidase alpha class 2A member 2 |
| chr15_+_65076744 | 1.42 |
ENST00000432196.5
|
KBTBD13
|
kelch repeat and BTB domain containing 13 |
| chrX_+_38561530 | 1.42 |
ENST00000378482.7
ENST00000286824.6 |
TSPAN7
|
tetraspanin 7 |
| chr16_+_83899079 | 1.41 |
ENST00000262430.6
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr9_-_33402449 | 1.28 |
ENST00000377425.8
|
AQP7
|
aquaporin 7 |
| chr14_-_72894091 | 1.26 |
ENST00000556509.6
|
DPF3
|
double PHD fingers 3 |
| chr3_-_58627596 | 1.23 |
ENST00000474531.5
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107 member A |
| chr8_+_95024977 | 1.19 |
ENST00000396124.9
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr15_-_43571497 | 1.18 |
ENST00000439195.5
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr5_+_1801387 | 1.16 |
ENST00000274137.10
ENST00000469176.1 |
NDUFS6
|
NADH:ubiquinone oxidoreductase subunit S6 |
| chr14_-_77320741 | 1.13 |
ENST00000682795.1
ENST00000682247.1 |
POMT2
|
protein O-mannosyltransferase 2 |
| chr8_+_12951583 | 1.09 |
ENST00000528753.2
|
TRMT9B
|
tRNA methyltransferase 9B (putative) |
| chr6_+_33204645 | 1.08 |
ENST00000374662.4
|
HSD17B8
|
hydroxysteroid 17-beta dehydrogenase 8 |
| chr6_+_159727561 | 1.08 |
ENST00000631126.2
ENST00000337387.4 |
WTAP
|
WT1 associated protein |
| chr16_-_88686453 | 1.03 |
ENST00000332281.6
|
SNAI3
|
snail family transcriptional repressor 3 |
| chr6_+_43770202 | 1.03 |
ENST00000372067.8
ENST00000672860.2 |
VEGFA
|
vascular endothelial growth factor A |
| chr3_-_58627567 | 1.01 |
ENST00000649301.1
|
FAM107A
|
family with sequence similarity 107 member A |
| chr4_+_158672237 | 0.98 |
ENST00000682734.1
|
ETFDH
|
electron transfer flavoprotein dehydrogenase |
| chr8_+_12951325 | 0.97 |
ENST00000400069.7
|
TRMT9B
|
tRNA methyltransferase 9B (putative) |
| chr9_-_33402551 | 0.97 |
ENST00000297988.6
ENST00000624075.3 ENST00000625032.1 ENST00000625109.3 |
AQP7
|
aquaporin 7 |
| chr19_-_50786139 | 0.96 |
ENST00000562076.2
|
ENSG00000261341.7
|
novel protein |
| chr12_-_124388811 | 0.90 |
ENST00000448614.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr10_-_61001430 | 0.90 |
ENST00000357917.4
|
RHOBTB1
|
Rho related BTB domain containing 1 |
| chr14_-_77320813 | 0.87 |
ENST00000682467.1
|
POMT2
|
protein O-mannosyltransferase 2 |
| chr19_-_43198079 | 0.86 |
ENST00000597374.5
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr22_-_38455199 | 0.86 |
ENST00000303592.3
|
KCNJ4
|
potassium inwardly rectifying channel subfamily J member 4 |
| chr14_-_21024092 | 0.82 |
ENST00000554398.5
|
NDRG2
|
NDRG family member 2 |
| chr2_-_213150236 | 0.81 |
ENST00000442445.1
ENST00000342002.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr12_-_53335737 | 0.81 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr20_-_45857196 | 0.80 |
ENST00000457981.5
ENST00000426915.1 ENST00000217455.9 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr14_-_77320855 | 0.79 |
ENST00000556394.2
ENST00000261534.9 |
POMT2
|
protein O-mannosyltransferase 2 |
| chr16_-_11915991 | 0.77 |
ENST00000420576.6
|
GSPT1
|
G1 to S phase transition 1 |
| chr6_+_31547560 | 0.76 |
ENST00000376148.9
ENST00000376145.8 |
NFKBIL1
|
NFKB inhibitor like 1 |
| chr11_-_26722051 | 0.76 |
ENST00000396005.8
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr14_+_73569115 | 0.75 |
ENST00000622407.4
ENST00000238651.10 |
ACOT2
|
acyl-CoA thioesterase 2 |
| chr22_+_29767351 | 0.74 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr12_-_53676072 | 0.74 |
ENST00000549748.2
ENST00000394349.9 |
ATP5MC2
|
ATP synthase membrane subunit c locus 2 |
| chr7_+_143052341 | 0.73 |
ENST00000418316.2
|
OR6V1
|
olfactory receptor family 6 subfamily V member 1 |
| chr1_+_61082398 | 0.72 |
ENST00000664149.1
|
NFIA
|
nuclear factor I A |
| chr3_+_150546765 | 0.70 |
ENST00000406576.7
ENST00000460851.6 ENST00000482093.5 ENST00000273435.9 |
EIF2A
|
eukaryotic translation initiation factor 2A |
| chr5_+_171420016 | 0.70 |
ENST00000675071.1
|
ENSG00000288639.1
|
novel protein |
| chr13_-_33185994 | 0.70 |
ENST00000255486.8
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr3_+_172126972 | 0.70 |
ENST00000443501.1
|
FNDC3B
|
fibronectin type III domain containing 3B |
| chr2_+_201071984 | 0.69 |
ENST00000237889.9
ENST00000433898.5 ENST00000684175.1 ENST00000682325.1 ENST00000454214.1 ENST00000684420.1 |
NDUFB3
|
NADH:ubiquinone oxidoreductase subunit B3 |
| chr12_-_53676314 | 0.69 |
ENST00000549164.5
|
ATP5MC2
|
ATP synthase membrane subunit c locus 2 |
| chr3_+_150546671 | 0.68 |
ENST00000487799.5
|
EIF2A
|
eukaryotic translation initiation factor 2A |
| chr14_+_73537135 | 0.67 |
ENST00000311148.9
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr6_+_159726998 | 0.66 |
ENST00000614346.4
|
WTAP
|
WT1 associated protein |
| chr8_-_80030232 | 0.66 |
ENST00000518271.1
ENST00000276585.9 ENST00000521605.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr15_-_82262660 | 0.62 |
ENST00000557844.1
ENST00000359445.7 ENST00000268206.12 |
EFL1
|
elongation factor like GTPase 1 |
| chr6_+_10585748 | 0.61 |
ENST00000265012.5
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr11_-_64245816 | 0.60 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1 regulatory inhibitor subunit 14B |
| chr1_+_215081793 | 0.58 |
ENST00000478774.5
|
KCNK2
|
potassium two pore domain channel subfamily K member 2 |
| chr4_-_185740211 | 0.58 |
ENST00000452351.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_-_108826161 | 0.56 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr19_+_50790415 | 0.55 |
ENST00000270593.2
|
ACP4
|
acid phosphatase 4 |
| chr1_-_54623518 | 0.55 |
ENST00000302250.7
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151 member A |
| chr16_-_11915878 | 0.55 |
ENST00000439887.6
ENST00000434724.7 |
GSPT1
|
G1 to S phase transition 1 |
| chr17_-_41586887 | 0.54 |
ENST00000167586.7
|
KRT14
|
keratin 14 |
| chr2_-_213150387 | 0.54 |
ENST00000433134.5
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr3_+_184186023 | 0.54 |
ENST00000429586.6
ENST00000292808.5 |
ABCF3
|
ATP binding cassette subfamily F member 3 |
| chr19_-_35812838 | 0.53 |
ENST00000653904.2
|
PRODH2
|
proline dehydrogenase 2 |
| chr2_-_201451446 | 0.53 |
ENST00000332624.8
ENST00000430254.1 |
TRAK2
|
trafficking kinesin protein 2 |
| chr14_+_77320996 | 0.52 |
ENST00000361389.8
ENST00000554279.5 ENST00000557639.5 ENST00000349555.7 ENST00000216465.10 ENST00000556627.5 ENST00000557053.5 |
GSTZ1
|
glutathione S-transferase zeta 1 |
| chr11_-_26721999 | 0.52 |
ENST00000280467.10
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr11_+_65530639 | 0.51 |
ENST00000279270.10
|
SCYL1
|
SCY1 like pseudokinase 1 |
| chr4_-_84499281 | 0.49 |
ENST00000295886.5
|
NKX6-1
|
NK6 homeobox 1 |
| chr22_-_31140494 | 0.48 |
ENST00000215885.4
|
PLA2G3
|
phospholipase A2 group III |
| chr6_+_54307856 | 0.48 |
ENST00000370869.7
|
TINAG
|
tubulointerstitial nephritis antigen |
| chr19_+_11239602 | 0.47 |
ENST00000252453.12
|
ANGPTL8
|
angiopoietin like 8 |
| chr20_+_19889502 | 0.47 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr8_-_80029904 | 0.46 |
ENST00000521434.5
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr1_+_44674728 | 0.45 |
ENST00000453711.1
|
ARMH1
|
armadillo like helical domain containing 1 |
| chr18_+_56651335 | 0.45 |
ENST00000589935.1
ENST00000254442.8 ENST00000357574.7 |
WDR7
|
WD repeat domain 7 |
| chr1_-_226186673 | 0.42 |
ENST00000366812.6
|
ACBD3
|
acyl-CoA binding domain containing 3 |
| chr2_+_108621260 | 0.41 |
ENST00000409441.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr14_-_21023954 | 0.41 |
ENST00000554094.5
|
NDRG2
|
NDRG family member 2 |
| chr1_-_145707345 | 0.40 |
ENST00000417171.6
|
PDZK1
|
PDZ domain containing 1 |
| chr19_-_4066892 | 0.39 |
ENST00000322357.9
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr19_-_35563653 | 0.36 |
ENST00000262623.4
|
ATP4A
|
ATPase H+/K+ transporting subunit alpha |
| chr12_-_42237727 | 0.36 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr3_-_126517764 | 0.34 |
ENST00000290868.7
|
UROC1
|
urocanate hydratase 1 |
| chr19_-_50968775 | 0.34 |
ENST00000391808.5
|
KLK6
|
kallikrein related peptidase 6 |
| chrX_-_50470818 | 0.34 |
ENST00000611977.2
|
DGKK
|
diacylglycerol kinase kappa |
| chr22_+_46762677 | 0.33 |
ENST00000355704.7
|
TBC1D22A
|
TBC1 domain family member 22A |
| chr2_+_209771972 | 0.33 |
ENST00000439458.5
ENST00000272845.10 |
UNC80
|
unc-80 homolog, NALCN channel complex subunit |
| chr22_+_46762617 | 0.33 |
ENST00000380995.5
ENST00000337137.9 ENST00000407381.7 |
TBC1D22A
|
TBC1 domain family member 22A |
| chr11_+_22668101 | 0.32 |
ENST00000630668.2
ENST00000278187.7 |
GAS2
|
growth arrest specific 2 |
| chr10_-_102056116 | 0.31 |
ENST00000370033.9
ENST00000311122.5 |
ARMH3
|
armadillo like helical domain containing 3 |
| chr14_+_21768482 | 0.31 |
ENST00000390428.3
|
TRAV6
|
T cell receptor alpha variable 6 |
| chr2_-_218269619 | 0.31 |
ENST00000447885.1
ENST00000420660.5 |
AAMP
|
angio associated migratory cell protein |
| chr2_+_33476640 | 0.29 |
ENST00000425210.5
ENST00000444784.5 ENST00000423159.5 ENST00000403687.8 |
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr1_-_22963178 | 0.29 |
ENST00000618559.1
|
LACTBL1
|
lactamase beta like 1 |
| chr12_-_118372883 | 0.29 |
ENST00000542532.5
ENST00000392533.8 |
TAOK3
|
TAO kinase 3 |
| chr8_-_80029826 | 0.29 |
ENST00000519386.5
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr13_+_21140514 | 0.28 |
ENST00000382533.8
ENST00000621421.4 |
SAP18
|
Sin3A associated protein 18 |
| chr3_-_113746059 | 0.27 |
ENST00000477813.5
|
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr1_+_61082702 | 0.26 |
ENST00000485903.6
ENST00000371185.6 ENST00000371184.6 |
NFIA
|
nuclear factor I A |
| chr2_-_218568291 | 0.26 |
ENST00000418019.5
ENST00000454775.5 ENST00000338465.5 ENST00000415516.5 ENST00000258399.8 |
USP37
|
ubiquitin specific peptidase 37 |
| chr1_-_145707387 | 0.25 |
ENST00000451928.6
|
PDZK1
|
PDZ domain containing 1 |
| chr11_-_134411854 | 0.25 |
ENST00000392580.5
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1 |
| chr11_-_64246190 | 0.25 |
ENST00000392210.6
|
PPP1R14B
|
protein phosphatase 1 regulatory inhibitor subunit 14B |
| chr3_-_113746218 | 0.24 |
ENST00000497255.1
ENST00000240922.8 ENST00000478020.1 ENST00000493900.5 |
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr11_-_65662419 | 0.23 |
ENST00000527874.1
|
RELA
|
RELA proto-oncogene, NF-kB subunit |
| chr2_+_74513441 | 0.23 |
ENST00000621092.1
|
TLX2
|
T cell leukemia homeobox 2 |
| chr3_-_126517745 | 0.23 |
ENST00000383579.3
|
UROC1
|
urocanate hydratase 1 |
| chr3_-_50567711 | 0.23 |
ENST00000357203.8
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr3_-_113746185 | 0.22 |
ENST00000616174.1
|
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr12_-_42238203 | 0.21 |
ENST00000327791.8
ENST00000534854.7 |
YAF2
|
YY1 associated factor 2 |
| chr11_-_62672255 | 0.21 |
ENST00000528862.2
|
LBHD1
|
LBH domain containing 1 |
| chr20_-_59032292 | 0.20 |
ENST00000395663.1
ENST00000243997.8 ENST00000395659.1 |
ATP5F1E
|
ATP synthase F1 subunit epsilon |
| chr1_-_19923617 | 0.20 |
ENST00000375116.3
|
PLA2G2E
|
phospholipase A2 group IIE |
| chr13_+_21140774 | 0.19 |
ENST00000450573.5
ENST00000467636.1 |
SAP18
|
Sin3A associated protein 18 |
| chr11_+_7573905 | 0.19 |
ENST00000529575.5
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr22_+_41381923 | 0.18 |
ENST00000266304.9
|
TEF
|
TEF transcription factor, PAR bZIP family member |
| chr19_-_40750302 | 0.17 |
ENST00000598485.6
ENST00000378313.7 ENST00000470681.5 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr1_-_44141631 | 0.17 |
ENST00000634670.1
|
KLF18
|
Kruppel like factor 18 |
| chr2_-_42361149 | 0.15 |
ENST00000468711.5
ENST00000463055.1 |
COX7A2L
|
cytochrome c oxidase subunit 7A2 like |
| chr17_-_41047267 | 0.15 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr11_-_73976952 | 0.14 |
ENST00000545212.1
|
UCP2
|
uncoupling protein 2 |
| chr4_-_75673112 | 0.14 |
ENST00000395719.7
ENST00000677489.1 |
G3BP2
|
G3BP stress granule assembly factor 2 |
| chr4_-_75673139 | 0.13 |
ENST00000677566.1
ENST00000503660.5 ENST00000677060.1 ENST00000678552.1 |
G3BP2
|
G3BP stress granule assembly factor 2 |
| chr1_-_27366917 | 0.13 |
ENST00000357582.3
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr20_+_48921701 | 0.13 |
ENST00000371917.5
|
ARFGEF2
|
ADP ribosylation factor guanine nucleotide exchange factor 2 |
| chr3_+_179347686 | 0.12 |
ENST00000471841.6
|
MFN1
|
mitofusin 1 |
| chr13_+_21140572 | 0.12 |
ENST00000607003.5
ENST00000492245.5 |
SAP18
|
Sin3A associated protein 18 |
| chr18_+_56651385 | 0.11 |
ENST00000615645.4
|
WDR7
|
WD repeat domain 7 |
| chr19_+_6464491 | 0.08 |
ENST00000308243.7
|
CRB3
|
crumbs cell polarity complex component 3 |
| chr6_-_142147122 | 0.07 |
ENST00000258042.2
|
NMBR
|
neuromedin B receptor |
| chrX_-_139204275 | 0.07 |
ENST00000441825.8
|
FGF13
|
fibroblast growth factor 13 |
| chr21_+_44012296 | 0.06 |
ENST00000291574.9
ENST00000380221.7 |
TRAPPC10
|
trafficking protein particle complex 10 |
| chr17_+_73193029 | 0.06 |
ENST00000299886.9
ENST00000438720.7 |
COG1
|
component of oligomeric golgi complex 1 |
| chr3_+_42856021 | 0.06 |
ENST00000493193.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr8_-_92103270 | 0.06 |
ENST00000518832.1
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr1_+_61082553 | 0.05 |
ENST00000403491.8
ENST00000371187.7 |
NFIA
|
nuclear factor I A |
| chr4_-_75672868 | 0.05 |
ENST00000678123.1
ENST00000678578.1 ENST00000677876.1 ENST00000676839.1 ENST00000678265.1 |
G3BP2
|
G3BP stress granule assembly factor 2 |
| chr10_-_71719772 | 0.01 |
ENST00000441508.4
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr11_-_72703385 | 0.01 |
ENST00000452383.2
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr4_-_98143416 | 0.01 |
ENST00000295268.4
|
STPG2
|
sperm tail PG-rich repeat containing 2 |
| chr12_+_51907478 | 0.01 |
ENST00000388922.9
|
ACVRL1
|
activin A receptor like type 1 |
| chr5_+_171419635 | 0.01 |
ENST00000274625.6
|
FGF18
|
fibroblast growth factor 18 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2C1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 12.0 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 1.8 | 9.0 | GO:0001315 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 1.2 | 3.7 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 1.1 | 8.7 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 1.1 | 3.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.9 | 2.7 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.8 | 4.8 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.8 | 5.5 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.5 | 3.9 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.4 | 1.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 2.8 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.3 | 1.0 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.2 | 3.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 0.7 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.2 | 2.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 0.6 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.2 | 0.9 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.2 | 1.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 1.4 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 4.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 5.5 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 0.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 2.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.5 | GO:0072560 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.1 | 1.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.6 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 2.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 2.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.5 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 | 0.6 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 2.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 4.8 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 1.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 2.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 1.2 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.7 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.8 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 1.7 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 3.6 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.2 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.5 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 3.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.8 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 2.0 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 1.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 1.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 1.7 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 1.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.7 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 1.8 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.5 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.4 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.3 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 6.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.4 | 1.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 4.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 2.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 15.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.3 | 1.4 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 1.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 3.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 3.1 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 2.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.6 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 4.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 3.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 1.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 9.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 3.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 4.3 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 10.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 3.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.9 | 9.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.8 | 2.5 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.5 | 5.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 4.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.5 | 2.0 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.4 | 4.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 2.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.4 | 2.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.4 | 1.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.3 | 2.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 1.5 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.3 | 0.8 | GO:0052816 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.2 | 0.7 | GO:0052858 | peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) |
| 0.2 | 3.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.2 | 2.4 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 3.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 1.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 1.7 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 1.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 12.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.4 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 1.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 1.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 4.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.7 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.7 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.6 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 6.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 2.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.8 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 2.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 1.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 2.5 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 11.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 7.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 3.4 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 4.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 12.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 9.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 4.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 3.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 4.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 3.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 4.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |