Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
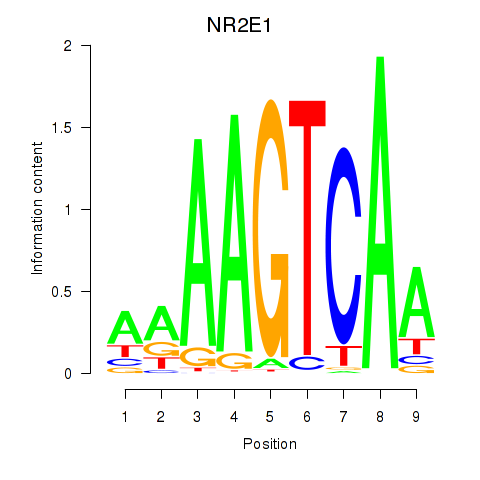
Results for NR2E1
Z-value: 0.69
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.12 | nuclear receptor subfamily 2 group E member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E1 | hg38_v1_chr6_+_108166015_108166034 | -0.43 | 1.4e-02 | Click! |
Activity profile of NR2E1 motif
Sorted Z-values of NR2E1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_116526277 | 2.09 |
ENST00000393468.1
ENST00000393467.1 |
CAV1
|
caveolin 1 |
| chr6_-_46954922 | 2.03 |
ENST00000265417.7
|
ADGRF5
|
adhesion G protein-coupled receptor F5 |
| chr3_-_64687613 | 1.64 |
ENST00000295903.8
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif 9 |
| chr1_+_185734362 | 1.63 |
ENST00000271588.9
|
HMCN1
|
hemicentin 1 |
| chr1_+_31576485 | 1.50 |
ENST00000457433.6
ENST00000271064.12 |
TINAGL1
|
tubulointerstitial nephritis antigen like 1 |
| chr3_-_64687992 | 1.46 |
ENST00000498707.5
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif 9 |
| chr2_+_191276885 | 1.43 |
ENST00000392316.5
|
MYO1B
|
myosin IB |
| chr2_-_89143133 | 1.41 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20 |
| chr11_-_5254741 | 1.40 |
ENST00000444587.1
ENST00000336906.6 ENST00000642908.1 ENST00000647543.1 |
HBG2
ENSG00000284931.1
|
hemoglobin subunit gamma 2 novel protein |
| chr3_-_64687982 | 1.28 |
ENST00000459780.1
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif 9 |
| chr5_-_33891941 | 1.26 |
ENST00000352040.7
ENST00000504830.6 |
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif 12 |
| chr7_+_143132069 | 1.20 |
ENST00000291009.4
|
PIP
|
prolactin induced protein |
| chr4_+_70197924 | 1.17 |
ENST00000514097.5
|
ODAM
|
odontogenic, ameloblast associated |
| chr1_-_94927079 | 1.12 |
ENST00000370206.9
ENST00000394202.8 |
CNN3
|
calponin 3 |
| chr5_-_33892013 | 1.11 |
ENST00000515401.1
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif 12 |
| chr17_-_48590231 | 1.11 |
ENST00000476342.1
ENST00000460160.5 ENST00000498678.6 ENST00000472863.5 |
HOXB3
|
homeobox B3 |
| chrX_-_66040072 | 1.11 |
ENST00000374737.9
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr5_-_91383310 | 1.09 |
ENST00000265138.4
|
ARRDC3
|
arrestin domain containing 3 |
| chr11_+_31812307 | 1.09 |
ENST00000643436.1
ENST00000646959.1 ENST00000645942.1 ENST00000530348.5 |
PAUPAR
ENSG00000285283.1
|
PAX6 upstream antisense RNA novel protein |
| chr12_-_91178520 | 1.08 |
ENST00000425043.5
ENST00000420120.6 ENST00000441303.6 ENST00000456569.2 |
DCN
|
decorin |
| chr8_-_121641424 | 1.08 |
ENST00000303924.5
|
HAS2
|
hyaluronan synthase 2 |
| chrX_-_66040057 | 1.06 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chrX_-_66040107 | 1.02 |
ENST00000455586.6
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr3_-_58210961 | 1.02 |
ENST00000486455.5
ENST00000394549.7 |
DNASE1L3
|
deoxyribonuclease 1 like 3 |
| chr5_+_32711313 | 1.01 |
ENST00000265074.13
|
NPR3
|
natriuretic peptide receptor 3 |
| chr11_-_5249836 | 1.00 |
ENST00000632727.1
ENST00000330597.5 |
HBG1
|
hemoglobin subunit gamma 1 |
| chr12_-_70637405 | 0.95 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr12_-_52493250 | 0.94 |
ENST00000330722.7
|
KRT6A
|
keratin 6A |
| chr5_+_32711723 | 0.94 |
ENST00000415167.2
|
NPR3
|
natriuretic peptide receptor 3 |
| chr13_+_110305806 | 0.92 |
ENST00000400163.7
|
COL4A2
|
collagen type IV alpha 2 chain |
| chr10_+_122560639 | 0.86 |
ENST00000344338.7
ENST00000330163.8 ENST00000652446.2 ENST00000666315.1 ENST00000368955.7 ENST00000368909.7 ENST00000368956.6 ENST00000619379.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr12_+_7062583 | 0.86 |
ENST00000542978.1
|
C1S
|
complement C1s |
| chr12_-_52949849 | 0.85 |
ENST00000619952.2
ENST00000546826.5 |
KRT8
|
keratin 8 |
| chr5_-_150137403 | 0.85 |
ENST00000517957.1
|
PDGFRB
|
platelet derived growth factor receptor beta |
| chrX_-_66033664 | 0.84 |
ENST00000427538.5
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr11_-_13495984 | 0.84 |
ENST00000282091.6
|
PTH
|
parathyroid hormone |
| chr6_+_31948956 | 0.83 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr6_+_148342759 | 0.83 |
ENST00000367467.8
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr18_+_44680093 | 0.82 |
ENST00000426838.8
ENST00000677068.1 |
SETBP1
|
SET binding protein 1 |
| chr9_-_92482499 | 0.79 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr11_-_13496018 | 0.79 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr3_+_12351470 | 0.78 |
ENST00000287820.10
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr6_-_41747390 | 0.78 |
ENST00000356667.8
ENST00000373025.7 ENST00000425343.6 |
PGC
|
progastricsin |
| chr11_+_102112445 | 0.77 |
ENST00000524575.5
|
YAP1
|
Yes1 associated transcriptional regulator |
| chr11_-_111923722 | 0.77 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B |
| chr9_-_92482350 | 0.76 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr9_-_92482461 | 0.75 |
ENST00000651738.1
|
ASPN
|
asporin |
| chr2_+_90038848 | 0.74 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr3_-_58211212 | 0.73 |
ENST00000461914.7
|
DNASE1L3
|
deoxyribonuclease 1 like 3 |
| chr19_-_46717076 | 0.73 |
ENST00000601806.5
ENST00000593363.1 ENST00000291281.9 ENST00000598633.1 ENST00000595515.5 ENST00000433867.5 |
PRKD2
|
protein kinase D2 |
| chr12_+_56521951 | 0.72 |
ENST00000552247.6
|
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr10_+_11742361 | 0.72 |
ENST00000379215.9
ENST00000420401.5 |
ECHDC3
|
enoyl-CoA hydratase domain containing 3 |
| chr3_-_112638097 | 0.72 |
ENST00000461431.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr17_-_68955332 | 0.71 |
ENST00000269080.6
ENST00000615593.4 ENST00000586539.6 ENST00000430352.6 |
ABCA8
|
ATP binding cassette subfamily A member 8 |
| chr10_+_122560751 | 0.71 |
ENST00000338354.10
ENST00000664692.1 ENST00000653442.1 ENST00000664974.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr16_+_3018390 | 0.71 |
ENST00000573001.5
|
TNFRSF12A
|
TNF receptor superfamily member 12A |
| chr12_+_56521990 | 0.71 |
ENST00000550726.5
ENST00000542360.1 |
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr5_-_42811884 | 0.69 |
ENST00000514985.6
ENST00000511224.5 ENST00000507920.5 ENST00000510965.1 |
SELENOP
|
selenoprotein P |
| chr4_-_184805752 | 0.68 |
ENST00000513317.5
|
ACSL1
|
acyl-CoA synthetase long chain family member 1 |
| chrX_+_47585212 | 0.68 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr2_-_88860913 | 0.67 |
ENST00000390239.2
|
IGKJ4
|
immunoglobulin kappa joining 4 |
| chr1_+_212565334 | 0.65 |
ENST00000366981.8
ENST00000366987.6 |
ATF3
|
activating transcription factor 3 |
| chr1_+_170664121 | 0.64 |
ENST00000239461.11
|
PRRX1
|
paired related homeobox 1 |
| chr3_+_12351493 | 0.64 |
ENST00000683699.1
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr20_-_31722854 | 0.63 |
ENST00000307677.5
|
BCL2L1
|
BCL2 like 1 |
| chr7_+_116672187 | 0.61 |
ENST00000318493.11
ENST00000397752.8 |
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr12_-_55842950 | 0.60 |
ENST00000548629.5
|
MMP19
|
matrix metallopeptidase 19 |
| chr9_+_1050330 | 0.60 |
ENST00000382255.7
ENST00000382251.7 ENST00000412350.6 ENST00000358146.7 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr20_-_31722533 | 0.59 |
ENST00000677194.1
ENST00000434194.2 ENST00000376062.6 |
BCL2L1
|
BCL2 like 1 |
| chr11_-_128587551 | 0.58 |
ENST00000392668.8
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr2_+_205682491 | 0.56 |
ENST00000360409.7
ENST00000450507.5 ENST00000357785.10 ENST00000417189.5 |
NRP2
|
neuropilin 2 |
| chr2_+_88897230 | 0.56 |
ENST00000390244.2
|
IGKV5-2
|
immunoglobulin kappa variable 5-2 |
| chrX_+_116170742 | 0.56 |
ENST00000371906.5
ENST00000681852.1 |
AGTR2
|
angiotensin II receptor type 2 |
| chr20_+_44531817 | 0.56 |
ENST00000372889.5
ENST00000372887.5 |
PKIG
|
cAMP-dependent protein kinase inhibitor gamma |
| chr20_-_31722949 | 0.55 |
ENST00000376055.9
|
BCL2L1
|
BCL2 like 1 |
| chr3_-_169146527 | 0.55 |
ENST00000475754.5
ENST00000484519.5 |
MECOM
|
MDS1 and EVI1 complex locus |
| chr1_+_170663917 | 0.53 |
ENST00000497230.2
|
PRRX1
|
paired related homeobox 1 |
| chr3_+_101827982 | 0.53 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr8_+_103372388 | 0.52 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr14_+_69398683 | 0.49 |
ENST00000556605.5
ENST00000031146.8 ENST00000336643.10 |
SLC39A9
|
solute carrier family 39 member 9 |
| chr6_+_142302621 | 0.49 |
ENST00000541199.5
ENST00000435011.6 |
ADGRG6
|
adhesion G protein-coupled receptor G6 |
| chr2_-_157444044 | 0.49 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein |
| chr12_+_85874287 | 0.48 |
ENST00000551529.5
ENST00000256010.7 |
NTS
|
neurotensin |
| chr18_+_44680875 | 0.48 |
ENST00000649279.2
ENST00000677699.1 |
SETBP1
|
SET binding protein 1 |
| chr3_-_50322733 | 0.47 |
ENST00000428028.1
ENST00000357750.9 |
HYAL2
|
hyaluronidase 2 |
| chr1_-_99766620 | 0.47 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1 |
| chr3_+_128052390 | 0.47 |
ENST00000481210.5
ENST00000243253.8 |
SEC61A1
|
SEC61 translocon subunit alpha 1 |
| chr3_-_169146595 | 0.47 |
ENST00000468789.5
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr10_+_122560679 | 0.47 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr15_-_51243011 | 0.46 |
ENST00000405913.7
ENST00000559878.5 |
CYP19A1
|
cytochrome P450 family 19 subfamily A member 1 |
| chr14_+_61697622 | 0.45 |
ENST00000539097.2
|
HIF1A
|
hypoxia inducible factor 1 subunit alpha |
| chr2_-_189580773 | 0.45 |
ENST00000261024.7
|
SLC40A1
|
solute carrier family 40 member 1 |
| chr3_-_50322759 | 0.44 |
ENST00000442581.1
ENST00000447092.5 |
HYAL2
|
hyaluronidase 2 |
| chr14_-_91836440 | 0.43 |
ENST00000340892.9
ENST00000360594.9 |
TC2N
|
tandem C2 domains, nuclear |
| chr6_-_138499487 | 0.43 |
ENST00000343505.9
|
NHSL1
|
NHS like 1 |
| chr8_+_24384455 | 0.43 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM like decysin 1 |
| chr8_+_24384275 | 0.43 |
ENST00000256412.8
|
ADAMDEC1
|
ADAM like decysin 1 |
| chr11_-_70717994 | 0.43 |
ENST00000659264.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr8_-_118111806 | 0.43 |
ENST00000378204.7
|
EXT1
|
exostosin glycosyltransferase 1 |
| chr11_-_111910830 | 0.42 |
ENST00000526167.5
ENST00000651650.1 |
CRYAB
|
crystallin alpha B |
| chr4_-_47981535 | 0.41 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr7_+_107168961 | 0.41 |
ENST00000468410.5
ENST00000478930.5 ENST00000464009.1 ENST00000222574.9 |
HBP1
|
HMG-box transcription factor 1 |
| chr3_+_136957948 | 0.41 |
ENST00000329582.9
|
IL20RB
|
interleukin 20 receptor subunit beta |
| chr17_-_43546323 | 0.41 |
ENST00000545954.5
ENST00000319349.10 |
ETV4
|
ETS variant transcription factor 4 |
| chr11_-_63671909 | 0.40 |
ENST00000538786.1
ENST00000540699.1 |
ATL3
|
atlastin GTPase 3 |
| chr6_-_138545685 | 0.40 |
ENST00000342260.9
|
NHSL1
|
NHS like 1 |
| chr12_-_91153149 | 0.40 |
ENST00000550758.1
|
DCN
|
decorin |
| chr12_-_31326142 | 0.40 |
ENST00000337682.9
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr12_-_55842927 | 0.40 |
ENST00000322569.9
ENST00000409200.7 |
MMP19
|
matrix metallopeptidase 19 |
| chr12_-_89352487 | 0.40 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr7_+_135148041 | 0.40 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr1_+_62597510 | 0.39 |
ENST00000371129.4
|
ANGPTL3
|
angiopoietin like 3 |
| chr14_+_36661852 | 0.39 |
ENST00000361487.7
|
PAX9
|
paired box 9 |
| chr9_-_72953047 | 0.39 |
ENST00000297785.8
ENST00000376939.5 |
ALDH1A1
|
aldehyde dehydrogenase 1 family member A1 |
| chr3_+_69739425 | 0.39 |
ENST00000352241.9
ENST00000642352.1 |
MITF
|
melanocyte inducing transcription factor |
| chr1_-_115768702 | 0.39 |
ENST00000261448.6
|
CASQ2
|
calsequestrin 2 |
| chr17_+_2056073 | 0.39 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
HIC ZBTB transcriptional repressor 1 |
| chrX_+_47218670 | 0.39 |
ENST00000357227.9
ENST00000519758.5 ENST00000520893.5 ENST00000622098.4 ENST00000517426.5 |
CDK16
|
cyclin dependent kinase 16 |
| chr6_+_158649997 | 0.38 |
ENST00000360448.8
ENST00000367081.7 ENST00000611299.5 |
SYTL3
|
synaptotagmin like 3 |
| chr6_+_143126183 | 0.38 |
ENST00000458219.1
|
AIG1
|
androgen induced 1 |
| chr12_+_63844758 | 0.38 |
ENST00000631006.2
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr12_+_10213417 | 0.38 |
ENST00000546017.5
ENST00000535576.5 ENST00000539170.5 |
GABARAPL1
|
GABA type A receptor associated protein like 1 |
| chr9_-_92404559 | 0.37 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chr4_+_77605807 | 0.36 |
ENST00000682537.1
|
CXCL13
|
C-X-C motif chemokine ligand 13 |
| chr11_-_111910790 | 0.36 |
ENST00000533280.6
|
CRYAB
|
crystallin alpha B |
| chr12_+_53990585 | 0.36 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr6_+_12290353 | 0.36 |
ENST00000379375.6
|
EDN1
|
endothelin 1 |
| chr1_-_243851066 | 0.36 |
ENST00000263826.12
|
AKT3
|
AKT serine/threonine kinase 3 |
| chr7_-_106285898 | 0.36 |
ENST00000424768.2
ENST00000681255.1 |
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr10_+_17752185 | 0.36 |
ENST00000377495.2
|
TMEM236
|
transmembrane protein 236 |
| chr3_+_52779916 | 0.36 |
ENST00000537050.5
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chrX_+_47218232 | 0.35 |
ENST00000457458.6
ENST00000522883.1 |
CDK16
|
cyclin dependent kinase 16 |
| chr21_-_34888683 | 0.35 |
ENST00000344691.8
ENST00000358356.9 |
RUNX1
|
RUNX family transcription factor 1 |
| chr2_+_74458400 | 0.35 |
ENST00000393972.7
ENST00000233615.7 ENST00000409737.5 ENST00000428943.1 |
WBP1
|
WW domain binding protein 1 |
| chr12_-_31326111 | 0.34 |
ENST00000539409.5
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr12_-_31326171 | 0.34 |
ENST00000542983.1
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr4_-_110198650 | 0.34 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr12_-_7936177 | 0.34 |
ENST00000544291.1
ENST00000075120.12 |
SLC2A3
|
solute carrier family 2 member 3 |
| chr1_-_243850216 | 0.34 |
ENST00000673466.1
|
AKT3
|
AKT serine/threonine kinase 3 |
| chr4_-_69639642 | 0.34 |
ENST00000604629.6
ENST00000604021.1 |
UGT2A2
|
UDP glucuronosyltransferase family 2 member A2 |
| chr7_-_42152396 | 0.34 |
ENST00000642432.1
ENST00000647255.1 ENST00000677288.1 |
GLI3
|
GLI family zinc finger 3 |
| chr14_-_91836526 | 0.33 |
ENST00000556018.5
|
TC2N
|
tandem C2 domains, nuclear |
| chr9_-_92536645 | 0.33 |
ENST00000395534.2
|
ECM2
|
extracellular matrix protein 2 |
| chr7_-_13986498 | 0.33 |
ENST00000420159.6
ENST00000399357.7 ENST00000403527.5 |
ETV1
|
ETS variant transcription factor 1 |
| chr12_+_10213624 | 0.32 |
ENST00000545290.1
|
GABARAPL1
|
GABA type A receptor associated protein like 1 |
| chr2_-_55010348 | 0.31 |
ENST00000394609.6
|
RTN4
|
reticulon 4 |
| chr20_+_43945677 | 0.31 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr4_-_40630588 | 0.31 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr17_+_75979211 | 0.30 |
ENST00000397640.6
ENST00000588202.5 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 subunit of CST complex |
| chr5_-_83673544 | 0.30 |
ENST00000503117.1
ENST00000510978.5 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr5_+_172834225 | 0.30 |
ENST00000393784.8
|
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment 1 |
| chr2_-_175181663 | 0.30 |
ENST00000392541.3
ENST00000284727.9 ENST00000409194.5 |
ATP5MC3
|
ATP synthase membrane subunit c locus 3 |
| chr7_-_16465728 | 0.30 |
ENST00000307068.5
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr7_+_114414997 | 0.29 |
ENST00000462331.5
ENST00000393491.7 ENST00000403559.8 ENST00000408937.7 ENST00000393498.6 ENST00000393495.7 ENST00000378237.7 |
FOXP2
|
forkhead box P2 |
| chr12_-_50249883 | 0.29 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr3_-_119677346 | 0.29 |
ENST00000484810.5
ENST00000497116.1 ENST00000261070.7 |
COX17
|
cytochrome c oxidase copper chaperone COX17 |
| chr19_+_35449584 | 0.29 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr8_-_18084925 | 0.29 |
ENST00000637792.1
ENST00000637991.1 ENST00000636537.1 ENST00000636455.1 ENST00000314146.10 ENST00000381733.9 |
ASAH1
|
N-acylsphingosine amidohydrolase 1 |
| chr4_-_40629842 | 0.29 |
ENST00000295971.12
|
RBM47
|
RNA binding motif protein 47 |
| chr3_+_69866217 | 0.29 |
ENST00000314589.10
|
MITF
|
melanocyte inducing transcription factor |
| chr11_-_104898670 | 0.29 |
ENST00000422698.6
|
CASP12
|
caspase 12 (gene/pseudogene) |
| chr16_-_69754913 | 0.29 |
ENST00000268802.10
|
NOB1
|
NIN1 (RPN12) binding protein 1 homolog |
| chr19_+_18001117 | 0.28 |
ENST00000379656.7
|
ARRDC2
|
arrestin domain containing 2 |
| chr17_-_69060906 | 0.28 |
ENST00000495634.5
ENST00000453985.6 ENST00000340001.9 ENST00000585714.1 |
ABCA9
|
ATP binding cassette subfamily A member 9 |
| chr2_-_189582012 | 0.28 |
ENST00000427419.5
ENST00000455320.5 |
SLC40A1
|
solute carrier family 40 member 1 |
| chr3_-_185821092 | 0.27 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr19_+_11435332 | 0.27 |
ENST00000591946.5
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr1_+_201739864 | 0.27 |
ENST00000367295.5
|
NAV1
|
neuron navigator 1 |
| chr4_-_110198579 | 0.27 |
ENST00000302274.8
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr12_-_57826295 | 0.27 |
ENST00000549039.5
|
CTDSP2
|
CTD small phosphatase 2 |
| chr3_+_122325237 | 0.26 |
ENST00000264474.4
ENST00000479204.1 |
CSTA
|
cystatin A |
| chr6_+_106098933 | 0.26 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr3_+_138621225 | 0.26 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_-_85578345 | 0.26 |
ENST00000426972.8
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr12_-_23949642 | 0.26 |
ENST00000537393.5
ENST00000451604.7 ENST00000381381.6 |
SOX5
|
SRY-box transcription factor 5 |
| chrX_-_49184789 | 0.26 |
ENST00000453382.5
ENST00000432913.5 |
PRICKLE3
|
prickle planar cell polarity protein 3 |
| chr12_-_29444236 | 0.26 |
ENST00000537054.1
|
OVCH1
|
ovochymase 1 |
| chr3_+_138621207 | 0.26 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr3_-_114178698 | 0.26 |
ENST00000467632.5
|
DRD3
|
dopamine receptor D3 |
| chr14_-_34874887 | 0.26 |
ENST00000382422.6
|
BAZ1A
|
bromodomain adjacent to zinc finger domain 1A |
| chr1_-_154183199 | 0.25 |
ENST00000323144.12
ENST00000330188.13 ENST00000328159.9 ENST00000611659.4 |
TPM3
|
tropomyosin 3 |
| chr9_-_92404690 | 0.25 |
ENST00000447356.1
|
OGN
|
osteoglycin |
| chr7_-_42152444 | 0.25 |
ENST00000479210.1
|
GLI3
|
GLI family zinc finger 3 |
| chr2_+_200308943 | 0.25 |
ENST00000619961.4
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr9_-_21994345 | 0.25 |
ENST00000579755.2
ENST00000530628.2 |
CDKN2A
|
cyclin dependent kinase inhibitor 2A |
| chr11_+_35176696 | 0.25 |
ENST00000528455.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr17_+_29568591 | 0.25 |
ENST00000581411.6
ENST00000301057.8 |
TP53I13
|
tumor protein p53 inducible protein 13 |
| chr11_+_35176639 | 0.24 |
ENST00000527889.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr7_-_113919000 | 0.24 |
ENST00000284601.4
|
PPP1R3A
|
protein phosphatase 1 regulatory subunit 3A |
| chr14_+_61187544 | 0.24 |
ENST00000555185.5
ENST00000557294.5 ENST00000556778.5 |
PRKCH
|
protein kinase C eta |
| chr12_+_119593758 | 0.23 |
ENST00000426426.3
|
TMEM233
|
transmembrane protein 233 |
| chr2_+_28395511 | 0.23 |
ENST00000436647.1
|
FOSL2
|
FOS like 2, AP-1 transcription factor subunit |
| chr15_+_67067780 | 0.23 |
ENST00000679624.1
|
SMAD3
|
SMAD family member 3 |
| chr11_+_35176611 | 0.23 |
ENST00000279452.10
|
CD44
|
CD44 molecule (Indian blood group) |
| chr14_+_51489112 | 0.22 |
ENST00000356218.8
|
FRMD6
|
FERM domain containing 6 |
| chr17_+_12666154 | 0.22 |
ENST00000343344.8
|
MYOCD
|
myocardin |
| chr5_-_16936231 | 0.22 |
ENST00000507288.1
ENST00000274203.13 ENST00000513610.6 |
MYO10
|
myosin X |
| chrM_+_10055 | 0.22 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 3 |
| chr11_+_35176575 | 0.22 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr7_+_116672357 | 0.22 |
ENST00000456159.1
|
MET
|
MET proto-oncogene, receptor tyrosine kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.4 | 2.4 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.3 | 2.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.3 | 1.6 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.3 | 2.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.3 | 0.8 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.2 | 4.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.7 | GO:0060345 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.2 | 0.9 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.2 | 1.8 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.2 | 4.4 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.2 | 0.6 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) negative regulation of icosanoid secretion(GO:0032304) |
| 0.2 | 1.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.2 | 0.4 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.2 | 1.4 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 2.1 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.2 | 0.7 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.6 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.6 | GO:0060366 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.1 | 2.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.1 | GO:0021546 | rhombomere development(GO:0021546) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.4 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 0.8 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 2.0 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 1.0 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 1.8 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.3 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.1 | 0.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.5 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.5 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.7 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.3 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.8 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 1.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.5 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 1.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.5 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.2 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 | 0.3 | GO:1904732 | regulation of electron carrier activity(GO:1904732) |
| 0.1 | 0.9 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 2.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.8 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 1.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.7 | GO:0089700 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) protein kinase D signaling(GO:0089700) |
| 0.1 | 0.9 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 0.3 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 | 0.8 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.2 | GO:1902688 | regulation of NAD metabolic process(GO:1902688) |
| 0.1 | 0.6 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.1 | 0.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) response to oleic acid(GO:0034201) |
| 0.1 | 0.2 | GO:0002665 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.1 | 1.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.3 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.6 | GO:1904996 | PML body organization(GO:0030578) positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.2 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.4 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.6 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.7 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.4 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.8 | GO:0072307 | contact inhibition(GO:0060242) metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 1.0 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 1.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.4 | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.0 | 0.1 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.2 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.2 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.5 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.0 | GO:0071650 | negative regulation of chemokine (C-C motif) ligand 5 production(GO:0071650) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.2 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 1.1 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:1900148 | Schwann cell proliferation involved in axon regeneration(GO:0014011) negative regulation of Schwann cell migration(GO:1900148) regulation of Schwann cell proliferation involved in axon regeneration(GO:1905044) negative regulation of Schwann cell proliferation involved in axon regeneration(GO:1905045) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.2 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.3 | GO:0043116 | arginine catabolic process(GO:0006527) negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.3 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.4 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 1.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0071753 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.2 | 0.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 0.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.4 | GO:0016590 | ACF complex(GO:0016590) |
| 0.1 | 1.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 2.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 1.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 2.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 2.0 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0097169 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.4 | GO:0033018 | junctional membrane complex(GO:0030314) sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:1990696 | stereocilia ankle link(GO:0002141) USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 12.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.5 | 2.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.3 | 2.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 0.7 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.2 | 1.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.8 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.2 | 0.9 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.4 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 1.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 2.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.6 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.4 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.5 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.2 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.7 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 7.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 5.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.4 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.8 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 1.1 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.0 | GO:0061598 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.2 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 3.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 3.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.3 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 9.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 3.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 2.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.8 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |