Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
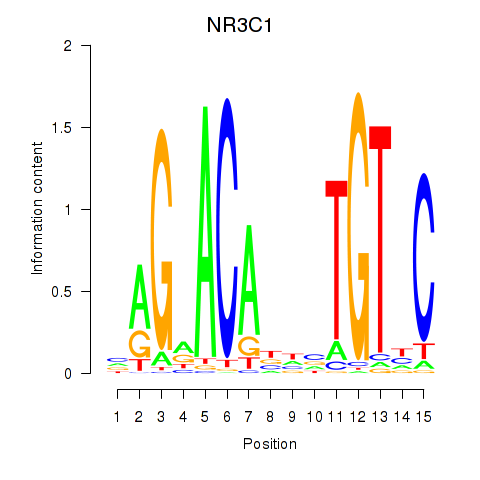
Results for NR3C1
Z-value: 2.23
Transcription factors associated with NR3C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR3C1
|
ENSG00000113580.15 | nuclear receptor subfamily 3 group C member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR3C1 | hg38_v1_chr5_-_143404536_143404667 | -0.32 | 7.3e-02 | Click! |
Activity profile of NR3C1 motif
Sorted Z-values of NR3C1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_22895368 | 11.41 |
ENST00000390321.2
|
IGLC1
|
immunoglobulin lambda constant 1 |
| chr14_-_105987068 | 10.76 |
ENST00000390594.3
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr22_+_22906342 | 10.74 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr22_+_22900976 | 9.79 |
ENST00000390323.2
|
IGLC2
|
immunoglobulin lambda constant 2 |
| chr14_-_105588322 | 8.83 |
ENST00000497872.4
ENST00000390539.2 |
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr2_-_89010515 | 8.58 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr2_-_89040745 | 7.58 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr15_-_21718245 | 7.23 |
ENST00000630556.1
|
ENSG00000281179.1
|
novel gene identicle to IGHV1OR15-1 |
| chr2_+_89959979 | 7.14 |
ENST00000453166.2
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr14_-_106235582 | 6.60 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr14_-_106130061 | 6.12 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr16_+_56632651 | 6.08 |
ENST00000379818.4
ENST00000570233.1 |
MT1M
|
metallothionein 1M |
| chr22_+_22380766 | 6.05 |
ENST00000390297.3
|
IGLV1-44
|
immunoglobulin lambda variable 1-44 |
| chr19_-_7732160 | 6.04 |
ENST00000676742.1
ENST00000678118.1 ENST00000328853.10 |
CLEC4G
|
C-type lectin domain family 4 member G |
| chr1_+_159587817 | 5.93 |
ENST00000255040.3
|
APCS
|
amyloid P component, serum |
| chr22_+_22357739 | 5.92 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr14_-_105708627 | 5.75 |
ENST00000641837.1
ENST00000390547.3 |
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr14_-_106349792 | 5.60 |
ENST00000438142.3
|
IGHV4-31
|
immunoglobulin heavy variable 4-31 |
| chr1_+_171090892 | 5.40 |
ENST00000367755.9
ENST00000479749.1 |
FMO3
|
flavin containing dimethylaniline monoxygenase 3 |
| chr2_-_89213917 | 5.37 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr2_-_136118142 | 5.26 |
ENST00000241393.4
|
CXCR4
|
C-X-C motif chemokine receptor 4 |
| chr16_+_32066065 | 5.22 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr22_+_22322452 | 5.21 |
ENST00000390290.3
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr12_-_46825949 | 5.15 |
ENST00000547477.5
ENST00000447411.5 ENST00000266579.9 |
SLC38A4
|
solute carrier family 38 member 4 |
| chr6_-_132714045 | 5.07 |
ENST00000367928.5
|
VNN1
|
vanin 1 |
| chr16_+_56682461 | 5.05 |
ENST00000562939.1
ENST00000394485.5 ENST00000567563.1 |
MT1X
ENSG00000259827.1
|
metallothionein 1X novel transcript |
| chr2_+_89936859 | 5.03 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr14_-_106538331 | 4.72 |
ENST00000390624.3
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr15_-_22160868 | 4.61 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr14_-_106117159 | 4.59 |
ENST00000390601.3
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 |
| chr16_+_33827140 | 4.53 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr14_-_106658251 | 4.51 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr2_-_89245596 | 4.48 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr14_-_106803221 | 4.33 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr15_-_89814845 | 4.29 |
ENST00000679248.1
ENST00000300060.7 ENST00000560137.2 |
ANPEP
|
alanyl aminopeptidase, membrane |
| chr22_+_22409755 | 4.23 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr22_+_22697789 | 4.19 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr14_-_106724093 | 4.14 |
ENST00000390634.3
|
IGHV2-70D
|
immunoglobulin heavy variable 2-70D |
| chr2_-_89268506 | 4.11 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr22_+_22792485 | 4.08 |
ENST00000390314.2
|
IGLV2-11
|
immunoglobulin lambda variable 2-11 |
| chr14_-_106511856 | 4.06 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr14_-_106675544 | 4.06 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr2_-_88128049 | 4.00 |
ENST00000393750.3
ENST00000295834.8 |
FABP1
|
fatty acid binding protein 1 |
| chr14_-_106811131 | 3.96 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr2_-_89222461 | 3.95 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr7_+_142352802 | 3.89 |
ENST00000634605.1
|
TRBV7-2
|
T cell receptor beta variable 7-2 |
| chr14_-_106422175 | 3.86 |
ENST00000390619.2
|
IGHV4-39
|
immunoglobulin heavy variable 4-39 |
| chr22_+_22327298 | 3.82 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr14_-_106012390 | 3.80 |
ENST00000455737.2
|
IGHV4-4
|
immunoglobulin heavy variable 4-4 |
| chr1_-_159714581 | 3.79 |
ENST00000255030.9
ENST00000437342.1 ENST00000368112.5 ENST00000368111.5 ENST00000368110.1 |
CRP
|
C-reactive protein |
| chr5_+_96876480 | 3.68 |
ENST00000437043.8
ENST00000379904.8 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr2_+_90154073 | 3.65 |
ENST00000611391.1
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr1_-_160523204 | 3.63 |
ENST00000368055.1
ENST00000368057.8 ENST00000368059.7 |
SLAMF6
|
SLAM family member 6 |
| chr19_-_10334723 | 3.60 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr15_+_88635626 | 3.51 |
ENST00000379224.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr1_-_206946448 | 3.50 |
ENST00000356495.5
|
PIGR
|
polymeric immunoglobulin receptor |
| chr14_-_106088573 | 3.38 |
ENST00000632099.1
|
IGHV3-64D
|
immunoglobulin heavy variable 3-64D |
| chr16_+_56625775 | 3.34 |
ENST00000330439.7
ENST00000568293.1 |
MT1E
|
metallothionein 1E |
| chr17_-_66229380 | 3.29 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr19_+_15640880 | 3.28 |
ENST00000586182.6
ENST00000221307.13 ENST00000591058.5 |
CYP4F3
|
cytochrome P450 family 4 subfamily F member 3 |
| chr8_+_11494367 | 3.26 |
ENST00000259089.9
ENST00000529894.1 |
BLK
|
BLK proto-oncogene, Src family tyrosine kinase |
| chr14_-_106737547 | 3.23 |
ENST00000632209.1
|
IGHV1-69-2
|
immunoglobulin heavy variable 1-69-2 |
| chr10_+_94938649 | 3.22 |
ENST00000461906.1
ENST00000260682.8 |
CYP2C9
|
cytochrome P450 family 2 subfamily C member 9 |
| chr12_+_95943318 | 3.15 |
ENST00000266736.7
|
AMDHD1
|
amidohydrolase domain containing 1 |
| chr3_-_42875871 | 3.13 |
ENST00000316161.6
ENST00000437102.1 |
CYP8B1
|
cytochrome P450 family 8 subfamily B member 1 |
| chr1_+_196819731 | 3.11 |
ENST00000320493.10
ENST00000367424.4 |
CFHR1
|
complement factor H related 1 |
| chr1_+_186296267 | 3.09 |
ENST00000533951.5
ENST00000367482.8 ENST00000635041.1 ENST00000367483.8 ENST00000367485.4 ENST00000445192.7 |
PRG4
|
proteoglycan 4 |
| chr3_+_186996444 | 3.04 |
ENST00000676633.1
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr14_-_106771020 | 3.02 |
ENST00000617374.2
|
IGHV2-70
|
immunoglobulin heavy variable 2-70 |
| chr22_+_22880706 | 3.02 |
ENST00000390319.2
|
IGLV3-1
|
immunoglobulin lambda variable 3-1 |
| chr16_+_56608577 | 2.99 |
ENST00000245185.6
ENST00000561491.1 |
MT2A
|
metallothionein 2A |
| chr1_-_1214146 | 2.98 |
ENST00000379236.4
|
TNFRSF4
|
TNF receptor superfamily member 4 |
| chr1_-_173917281 | 2.95 |
ENST00000367698.4
|
SERPINC1
|
serpin family C member 1 |
| chr16_+_30472733 | 2.94 |
ENST00000356798.11
ENST00000433423.2 |
ITGAL
|
integrin subunit alpha L |
| chr14_-_106301848 | 2.92 |
ENST00000390611.2
|
IGHV2-26
|
immunoglobulin heavy variable 2-26 |
| chr14_-_106154113 | 2.90 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr1_+_207089233 | 2.88 |
ENST00000243611.9
ENST00000367076.7 |
C4BPB
|
complement component 4 binding protein beta |
| chr4_+_22692906 | 2.86 |
ENST00000613293.4
ENST00000610628.4 |
GBA3
|
glucosylceramidase beta 3 (gene/pseudogene) |
| chr3_+_52779916 | 2.82 |
ENST00000537050.5
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr1_-_56966133 | 2.78 |
ENST00000535057.5
ENST00000543257.5 |
C8B
|
complement C8 beta chain |
| chr11_+_117986386 | 2.73 |
ENST00000227752.8
|
IL10RA
|
interleukin 10 receptor subunit alpha |
| chr3_+_52787825 | 2.73 |
ENST00000405128.3
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr22_+_44180915 | 2.71 |
ENST00000444313.8
ENST00000416291.5 |
PARVG
|
parvin gamma |
| chr9_-_37034031 | 2.69 |
ENST00000520281.5
ENST00000446742.5 ENST00000522003.5 ENST00000523145.5 ENST00000414447.5 ENST00000377847.6 ENST00000377853.6 |
PAX5
|
paired box 5 |
| chr1_+_207089195 | 2.69 |
ENST00000452902.6
|
C4BPB
|
complement component 4 binding protein beta |
| chr2_+_181457342 | 2.68 |
ENST00000397033.7
ENST00000233573.6 |
ITGA4
|
integrin subunit alpha 4 |
| chr17_-_39864140 | 2.66 |
ENST00000623724.3
ENST00000439167.6 ENST00000377945.7 ENST00000394189.6 ENST00000377944.7 ENST00000377958.6 ENST00000535189.5 ENST00000377952.6 |
IKZF3
|
IKAROS family zinc finger 3 |
| chr2_-_88861563 | 2.64 |
ENST00000624935.3
ENST00000390241.3 |
ENSG00000240040.6
IGKJ2
|
novel transcript immunoglobulin kappa joining 2 |
| chr22_+_22922594 | 2.61 |
ENST00000390331.3
|
IGLC7
|
immunoglobulin lambda constant 7 |
| chr14_-_106211453 | 2.58 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr14_-_106324743 | 2.52 |
ENST00000390612.3
|
IGHV4-28
|
immunoglobulin heavy variable 4-28 |
| chr11_+_57597563 | 2.50 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chr1_+_207089283 | 2.49 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein beta |
| chr3_-_151329539 | 2.49 |
ENST00000325602.6
|
P2RY13
|
purinergic receptor P2Y13 |
| chr16_+_3065380 | 2.48 |
ENST00000551122.5
ENST00000548807.5 ENST00000528163.6 |
IL32
|
interleukin 32 |
| chr1_-_206921867 | 2.44 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr19_-_15898057 | 2.42 |
ENST00000011989.11
ENST00000221700.11 |
CYP4F2
|
cytochrome P450 family 4 subfamily F member 2 |
| chr1_-_11047225 | 2.42 |
ENST00000400898.3
ENST00000400897.8 |
MASP2
|
mannan binding lectin serine peptidase 2 |
| chr1_+_207088825 | 2.40 |
ENST00000367078.8
|
C4BPB
|
complement component 4 binding protein beta |
| chr20_+_57561103 | 2.39 |
ENST00000319441.6
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 |
| chr10_-_50885656 | 2.37 |
ENST00000374001.6
ENST00000395489.6 ENST00000282641.6 ENST00000395495.5 ENST00000373995.7 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr2_+_241809179 | 2.36 |
ENST00000405370.5
ENST00000407683.6 |
NEU4
|
neuraminidase 4 |
| chr16_+_56638659 | 2.34 |
ENST00000290705.12
|
MT1A
|
metallothionein 1A |
| chr21_-_32813679 | 2.31 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr10_+_4963406 | 2.31 |
ENST00000380872.9
ENST00000442997.5 |
AKR1C1
|
aldo-keto reductase family 1 member C1 |
| chr17_-_82317523 | 2.29 |
ENST00000583376.1
ENST00000578509.1 ENST00000584284.5 ENST00000582480.1 |
CD7
|
CD7 molecule |
| chr4_+_73481737 | 2.28 |
ENST00000226355.5
|
AFM
|
afamin |
| chr11_+_114296347 | 2.26 |
ENST00000299964.4
|
NNMT
|
nicotinamide N-methyltransferase |
| chr16_+_31355215 | 2.25 |
ENST00000562522.2
|
ITGAX
|
integrin subunit alpha X |
| chr14_-_106005574 | 2.24 |
ENST00000390595.3
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr3_+_187024614 | 2.21 |
ENST00000416235.6
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr12_-_121858849 | 2.21 |
ENST00000289004.8
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chr1_-_206921987 | 2.19 |
ENST00000530505.1
ENST00000442471.4 |
FCMR
|
Fc fragment of IgM receptor |
| chr4_+_15778320 | 2.19 |
ENST00000226279.8
|
CD38
|
CD38 molecule |
| chr3_+_122325237 | 2.17 |
ENST00000264474.4
ENST00000479204.1 |
CSTA
|
cystatin A |
| chr17_+_75979211 | 2.15 |
ENST00000397640.6
ENST00000588202.5 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 subunit of CST complex |
| chr10_+_69088096 | 2.13 |
ENST00000242465.4
|
SRGN
|
serglycin |
| chr2_+_113406368 | 2.13 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr17_-_3696198 | 2.13 |
ENST00000345901.7
|
P2RX5
|
purinergic receptor P2X 5 |
| chr16_+_82035245 | 2.13 |
ENST00000199936.9
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr2_-_157444044 | 2.12 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein |
| chr2_+_233729042 | 2.11 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chr1_-_206923242 | 2.11 |
ENST00000529560.1
|
FCMR
|
Fc fragment of IgM receptor |
| chr19_-_6670117 | 2.11 |
ENST00000245912.7
|
TNFSF14
|
TNF superfamily member 14 |
| chr14_-_106593319 | 2.10 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr4_-_99352754 | 2.09 |
ENST00000639454.1
|
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr16_+_3065311 | 2.09 |
ENST00000534507.5
ENST00000613483.4 ENST00000531965.5 ENST00000396887.7 |
IL32
|
interleukin 32 |
| chr14_-_106411021 | 2.08 |
ENST00000390618.2
|
IGHV3-38
|
immunoglobulin heavy variable 3-38 (non-functional) |
| chr16_-_10559135 | 2.04 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr14_+_21852457 | 2.04 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3 |
| chr2_-_75561297 | 2.04 |
ENST00000410071.5
ENST00000432649.5 |
EVA1A
|
eva-1 homolog A, regulator of programmed cell death |
| chr7_+_142300924 | 2.02 |
ENST00000455382.2
|
TRBV2
|
T cell receptor beta variable 2 |
| chr22_+_22771791 | 2.01 |
ENST00000390313.3
|
IGLV3-12
|
immunoglobulin lambda variable 3-12 |
| chr16_+_31355165 | 2.00 |
ENST00000562918.5
ENST00000268296.9 |
ITGAX
|
integrin subunit alpha X |
| chr5_+_134114673 | 2.00 |
ENST00000342854.10
ENST00000395029.5 |
TCF7
|
transcription factor 7 |
| chr3_-_119559599 | 1.99 |
ENST00000264246.8
|
CD80
|
CD80 molecule |
| chr15_-_78944985 | 1.99 |
ENST00000615999.5
ENST00000677789.1 ENST00000676880.1 ENST00000677936.1 ENST00000220166.10 ENST00000677810.1 ENST00000678644.1 ENST00000677534.1 ENST00000677316.1 |
CTSH
|
cathepsin H |
| chr16_+_56651885 | 1.96 |
ENST00000334346.3
ENST00000562399.1 |
MT1B
|
metallothionein 1B |
| chr15_-_79896985 | 1.96 |
ENST00000258874.4
|
MTHFS
|
methenyltetrahydrofolate synthetase |
| chr17_-_3696133 | 1.95 |
ENST00000225328.10
|
P2RX5
|
purinergic receptor P2X 5 |
| chr21_-_32813695 | 1.93 |
ENST00000479548.2
ENST00000490358.5 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr19_+_15641280 | 1.92 |
ENST00000585846.1
|
CYP4F3
|
cytochrome P450 family 4 subfamily F member 3 |
| chr3_+_46370854 | 1.92 |
ENST00000292303.4
|
CCR5
|
C-C motif chemokine receptor 5 |
| chr16_+_30472700 | 1.91 |
ENST00000358164.9
|
ITGAL
|
integrin subunit alpha L |
| chr9_-_37034185 | 1.90 |
ENST00000520154.6
|
PAX5
|
paired box 5 |
| chr19_-_51417581 | 1.90 |
ENST00000442846.7
ENST00000530476.1 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr4_-_99352730 | 1.89 |
ENST00000510055.5
ENST00000515683.6 ENST00000511397.3 |
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide |
| chr17_-_3696033 | 1.89 |
ENST00000551178.5
ENST00000552276.5 ENST00000547178.5 |
P2RX5
|
purinergic receptor P2X 5 |
| chr22_+_44181357 | 1.88 |
ENST00000417767.1
|
PARVG
|
parvin gamma |
| chr9_+_114155526 | 1.88 |
ENST00000356083.8
|
COL27A1
|
collagen type XXVII alpha 1 chain |
| chr16_+_33009175 | 1.87 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr20_-_20712626 | 1.87 |
ENST00000202677.12
|
RALGAPA2
|
Ral GTPase activating protein catalytic subunit alpha 2 |
| chr14_-_105864247 | 1.86 |
ENST00000461719.1
|
IGHJ4
|
immunoglobulin heavy joining 4 |
| chr3_+_52211442 | 1.85 |
ENST00000459884.1
|
ALAS1
|
5'-aminolevulinate synthase 1 |
| chr14_-_106470788 | 1.83 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr15_+_32717994 | 1.83 |
ENST00000560677.5
ENST00000560830.1 ENST00000651154.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr3_+_186640355 | 1.82 |
ENST00000382134.7
ENST00000265029.8 |
FETUB
|
fetuin B |
| chr12_-_7091873 | 1.81 |
ENST00000538050.5
ENST00000536053.6 |
C1R
|
complement C1r |
| chr10_-_52771700 | 1.77 |
ENST00000373968.3
|
MBL2
|
mannose binding lectin 2 |
| chr4_-_152679984 | 1.76 |
ENST00000304385.8
ENST00000504064.1 |
TMEM154
|
transmembrane protein 154 |
| chr10_-_133276855 | 1.76 |
ENST00000486609.1
ENST00000445355.8 ENST00000485491.6 |
ADAM8
|
ADAM metallopeptidase domain 8 |
| chr15_+_81182579 | 1.74 |
ENST00000302987.9
|
IL16
|
interleukin 16 |
| chr12_-_95995920 | 1.74 |
ENST00000552509.5
|
HAL
|
histidine ammonia-lyase |
| chr9_-_94640130 | 1.73 |
ENST00000414122.1
|
FBP1
|
fructose-bisphosphatase 1 |
| chr1_-_197067234 | 1.73 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr2_+_130356036 | 1.72 |
ENST00000347849.7
ENST00000175756.10 |
PTPN18
|
protein tyrosine phosphatase non-receptor type 18 |
| chr1_-_56966006 | 1.72 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain |
| chr19_+_13150386 | 1.70 |
ENST00000292433.4
ENST00000587885.1 |
IER2
|
immediate early response 2 |
| chr7_+_139778229 | 1.67 |
ENST00000425687.5
ENST00000650822.1 ENST00000416849.6 ENST00000438104.6 ENST00000336425.10 |
TBXAS1
|
thromboxane A synthase 1 |
| chr19_+_49335396 | 1.67 |
ENST00000598095.5
ENST00000426897.6 ENST00000323906.9 ENST00000535669.6 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr9_+_4839761 | 1.65 |
ENST00000448872.6
ENST00000441844.2 |
RCL1
|
RNA terminal phosphate cyclase like 1 |
| chr10_-_50885619 | 1.65 |
ENST00000373997.8
|
A1CF
|
APOBEC1 complementation factor |
| chr10_-_133276836 | 1.64 |
ENST00000415217.7
|
ADAM8
|
ADAM metallopeptidase domain 8 |
| chr7_+_142308517 | 1.63 |
ENST00000390387.3
|
TRBV3-1
|
T cell receptor beta variable 3-1 |
| chr17_-_75765136 | 1.63 |
ENST00000592997.6
ENST00000588479.6 ENST00000225614.6 |
GALK1
|
galactokinase 1 |
| chr15_-_55408467 | 1.62 |
ENST00000310958.10
|
CCPG1
|
cell cycle progression 1 |
| chr4_+_140524179 | 1.62 |
ENST00000507667.1
|
ELMOD2
|
ELMO domain containing 2 |
| chr19_-_10339610 | 1.60 |
ENST00000589261.5
ENST00000160262.10 ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr6_+_32038382 | 1.59 |
ENST00000478281.5
ENST00000471671.4 ENST00000435122.3 ENST00000644719.2 |
CYP21A2
|
cytochrome P450 family 21 subfamily A member 2 |
| chr19_-_51417619 | 1.59 |
ENST00000441969.7
ENST00000339313.10 ENST00000525998.5 ENST00000436984.6 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr16_+_71526419 | 1.59 |
ENST00000539698.4
|
CHST4
|
carbohydrate sulfotransferase 4 |
| chr18_+_49561013 | 1.59 |
ENST00000583083.1
|
LIPG
|
lipase G, endothelial type |
| chr4_+_68815991 | 1.58 |
ENST00000265403.12
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase family 2 member B10 |
| chr7_+_101218146 | 1.58 |
ENST00000305105.3
|
ZNHIT1
|
zinc finger HIT-type containing 1 |
| chr1_-_150765735 | 1.57 |
ENST00000679898.1
ENST00000448301.7 ENST00000680664.1 ENST00000679512.1 ENST00000368985.8 ENST00000679582.1 |
CTSS
|
cathepsin S |
| chr17_+_7583828 | 1.55 |
ENST00000396501.8
ENST00000250124.11 ENST00000584378.5 ENST00000423172.6 ENST00000579445.5 ENST00000585217.5 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr15_+_31366138 | 1.54 |
ENST00000558844.1
|
KLF13
|
Kruppel like factor 13 |
| chr14_+_22147988 | 1.54 |
ENST00000390457.2
|
TRAV27
|
T cell receptor alpha variable 27 |
| chr3_+_128052390 | 1.54 |
ENST00000481210.5
ENST00000243253.8 |
SEC61A1
|
SEC61 translocon subunit alpha 1 |
| chr17_-_3691748 | 1.53 |
ENST00000552723.1
|
P2RX5
|
purinergic receptor P2X 5 |
| chr19_-_58353482 | 1.52 |
ENST00000263100.8
|
A1BG
|
alpha-1-B glycoprotein |
| chr2_+_233712905 | 1.52 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr12_-_7092529 | 1.51 |
ENST00000540610.5
|
C1R
|
complement C1r |
| chr16_+_3065297 | 1.50 |
ENST00000325568.9
|
IL32
|
interleukin 32 |
| chr17_-_5584448 | 1.48 |
ENST00000269280.8
ENST00000571451.6 ENST00000572272.6 ENST00000613500.4 ENST00000619223.4 ENST00000617618.4 ENST00000345221.7 ENST00000262467.10 |
NLRP1
|
NLR family pyrin domain containing 1 |
| chr17_+_54938848 | 1.48 |
ENST00000574318.1
|
TOM1L1
|
target of myb1 like 1 membrane trafficking protein |
| chr19_-_55325316 | 1.48 |
ENST00000591570.5
ENST00000326652.9 |
TMEM150B
|
transmembrane protein 150B |
| chr6_-_26234978 | 1.47 |
ENST00000244534.7
|
H1-3
|
H1.3 linker histone, cluster member |
| chr19_+_16197900 | 1.47 |
ENST00000429941.6
ENST00000291439.8 ENST00000444449.6 ENST00000589822.5 |
AP1M1
|
adaptor related protein complex 1 subunit mu 1 |
| chr15_+_58410543 | 1.47 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
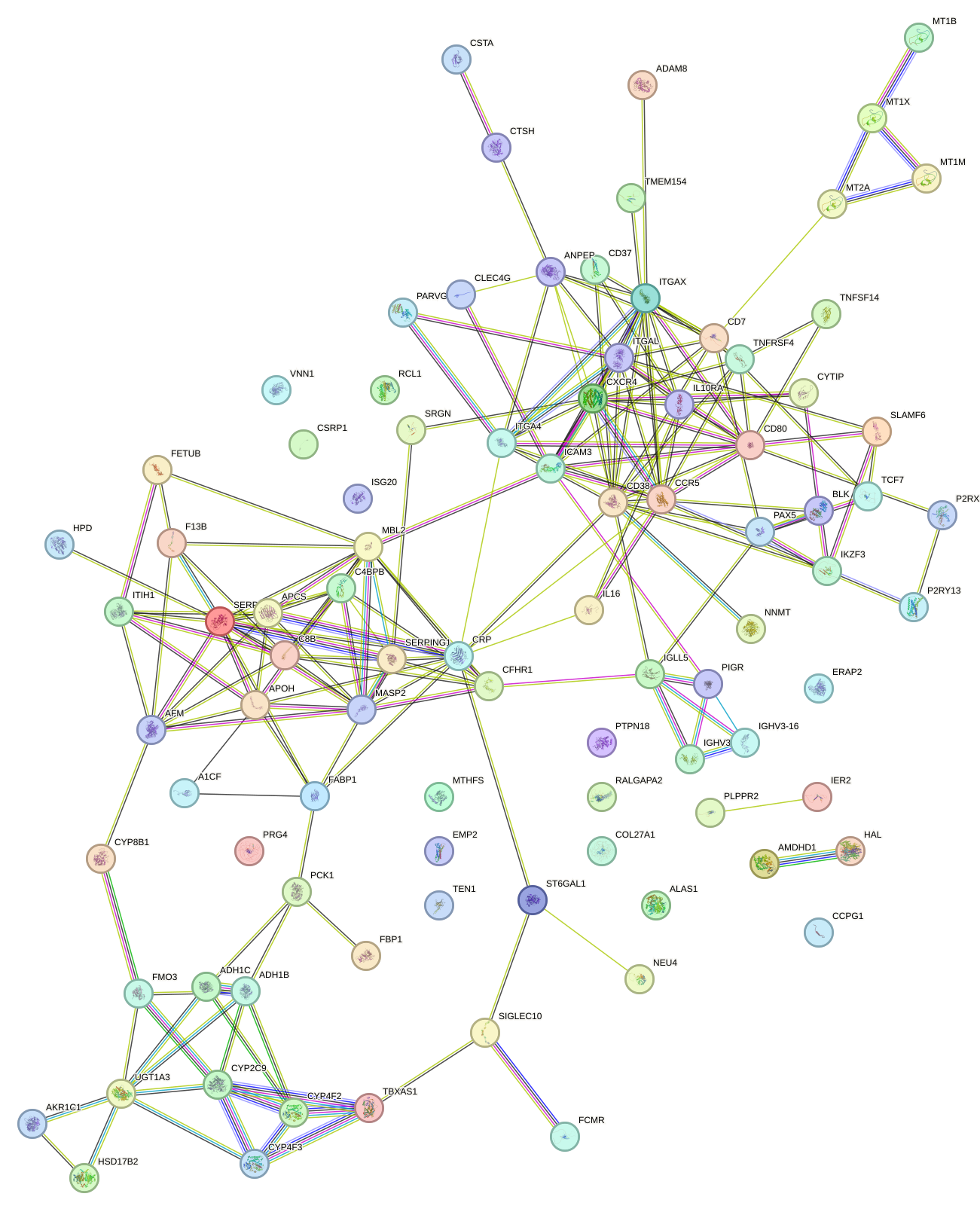
Network of associatons between targets according to the STRING database.
First level regulatory network of NR3C1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 1.4 | 8.3 | GO:1901569 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 1.4 | 231.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 1.2 | 3.5 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.1 | 8.0 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 1.1 | 3.4 | GO:2000309 | positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) positive regulation of thymocyte migration(GO:2000412) |
| 1.0 | 2.9 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.9 | 3.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.8 | 4.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.8 | 9.7 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.8 | 2.4 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.8 | 2.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.7 | 4.4 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.7 | 2.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.7 | 4.9 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.7 | 3.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.7 | 6.7 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.6 | 2.5 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.6 | 1.8 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.6 | 2.3 | GO:0050822 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.5 | 2.1 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.5 | 1.6 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.5 | 5.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.5 | 2.0 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.5 | 1.5 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.5 | 1.9 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.5 | 3.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.4 | 2.7 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.4 | 5.2 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.4 | 1.7 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.4 | 12.9 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.4 | 3.0 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.4 | 1.3 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) negative regulation of interleukin-3 production(GO:0032712) negative regulation of granulocyte colony-stimulating factor production(GO:0071656) negative regulation of macrophage colony-stimulating factor production(GO:1901257) |
| 0.4 | 0.8 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.4 | 0.8 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.4 | 1.2 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.4 | 0.8 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.4 | 2.0 | GO:0015942 | formate metabolic process(GO:0015942) |
| 0.4 | 1.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.4 | 1.1 | GO:0010931 | macrophage tolerance induction(GO:0010931) regulation of macrophage tolerance induction(GO:0010932) positive regulation of macrophage tolerance induction(GO:0010933) |
| 0.4 | 1.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.4 | 3.2 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.3 | 4.8 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.3 | 3.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 4.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 1.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.3 | 1.3 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.3 | 4.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 4.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.3 | 1.3 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.3 | 1.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.3 | 1.9 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.3 | 1.5 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.3 | 5.5 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.3 | 6.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.3 | 1.4 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 | 1.1 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.3 | 1.4 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.3 | 4.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.3 | 0.8 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.3 | 1.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.3 | 0.8 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.3 | 0.5 | GO:1904640 | response to methionine(GO:1904640) |
| 0.3 | 10.3 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.2 | 2.0 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 | 5.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 4.7 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.2 | 3.5 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.2 | 1.2 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 1.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.2 | 1.5 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.2 | 1.1 | GO:0038156 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.2 | 3.9 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.2 | 0.9 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 0.6 | GO:0097536 | thymus epithelium morphogenesis(GO:0097536) |
| 0.2 | 1.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.2 | 0.8 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 1.7 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.2 | 0.8 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.2 | 2.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 0.8 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.2 | 9.7 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.2 | 1.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 4.0 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 5.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 5.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 0.6 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.2 | 3.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 0.4 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 0.2 | 1.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 1.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 0.7 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 0.4 | GO:0035419 | activation of MAPK activity involved in innate immune response(GO:0035419) |
| 0.2 | 0.5 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.2 | 0.9 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.2 | 0.7 | GO:0032595 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.2 | 1.0 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 0.2 | 1.4 | GO:2000230 | response to metformin(GO:1901558) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 0.9 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.2 | 4.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.2 | 2.0 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.2 | 1.0 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.2 | 0.8 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 0.3 | GO:0019230 | proprioception(GO:0019230) |
| 0.2 | 0.5 | GO:0071626 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.2 | 1.0 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.2 | 0.6 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.2 | 0.9 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.2 | 1.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.6 | GO:2001301 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.2 | 2.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.6 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.1 | 1.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.7 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.1 | 1.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.7 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.7 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 1.0 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.7 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 2.0 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.5 | GO:0018277 | protein deamination(GO:0018277) |
| 0.1 | 0.5 | GO:0050992 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.1 | 0.9 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.7 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 0.4 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 1.0 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 1.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 1.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.6 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 2.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.4 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 26.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.8 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 1.7 | GO:0002423 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.1 | 0.7 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.7 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 1.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 1.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.1 | 0.3 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.1 | 0.7 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 3.0 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.6 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 1.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 1.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.4 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.1 | 1.0 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.1 | 1.4 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.5 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.1 | 2.7 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.1 | 1.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 1.8 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 | 0.3 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.1 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.1 | 0.3 | GO:0100057 | regulation of phenotypic switching by transcription from RNA polymerase II promoter(GO:0100057) regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.1 | 0.9 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 0.7 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 0.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.1 | 1.7 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 8.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.6 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 1.8 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 | 0.7 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 1.5 | GO:0034638 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 1.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 34.9 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 0.4 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.1 | 0.4 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.1 | 1.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.1 | 2.3 | GO:0043485 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 0.2 | GO:0044467 | glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.1 | 2.0 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 5.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.6 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 1.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.4 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.1 | 4.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.7 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.5 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.1 | 0.9 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.1 | 1.1 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 1.9 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.8 | GO:0042346 | positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.1 | 1.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.4 | GO:0031337 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.1 | 0.4 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.1 | GO:0072573 | propionate metabolic process(GO:0019541) negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 0.3 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.1 | 0.3 | GO:0015744 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 1.0 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 1.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 2.3 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 1.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.6 | GO:0015827 | L-alanine transport(GO:0015808) tryptophan transport(GO:0015827) |
| 0.1 | 0.2 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.1 | 0.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.8 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 1.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.3 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.1 | 0.5 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 1.0 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.8 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 0.9 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.4 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.5 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.4 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 2.6 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 1.4 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.0 | 0.7 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.2 | GO:0003069 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.2 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.0 | 0.5 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 1.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 3.6 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 1.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 1.0 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.2 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.4 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.8 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.6 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 1.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.6 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 5.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 1.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.3 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.4 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.6 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 1.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 6.1 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.9 | GO:0072678 | T cell migration(GO:0072678) |
| 0.0 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 1.4 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 1.1 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.3 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.9 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.8 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.8 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 1.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.6 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 4.6 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 1.9 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.6 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.4 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 1.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.4 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.3 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 1.5 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.9 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.9 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 2.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.7 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.5 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.4 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 2.7 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 1.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.4 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.4 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.9 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0033081 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.4 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.0 | 0.2 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 1.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.4 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 4.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 2.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.0 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.6 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:0090299 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.0 | GO:0010446 | response to alkaline pH(GO:0010446) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 1.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 1.4 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 1.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.0 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.2 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.6 | GO:0071750 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) |
| 1.6 | 85.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.1 | 3.4 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 1.0 | 4.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.8 | 4.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.6 | 6.3 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.6 | 2.3 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.5 | 3.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.5 | 2.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.5 | 4.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 2.4 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.4 | 1.2 | GO:1990696 | stereocilia ankle link(GO:0002141) USH2 complex(GO:1990696) |
| 0.4 | 2.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.4 | 1.1 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 0.3 | 71.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 2.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.3 | 0.8 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.3 | 4.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 7.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.2 | 1.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 4.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 0.8 | GO:0097196 | Shu complex(GO:0097196) |
| 0.2 | 1.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 1.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 1.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 2.3 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.1 | 0.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 7.2 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.4 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 0.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 11.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 1.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.7 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.4 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.8 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 2.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 2.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.3 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 1.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 6.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.9 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.3 | GO:0070701 | mucus layer(GO:0070701) |
| 0.1 | 2.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 9.6 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 0.3 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 0.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.6 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 7.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 4.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 1.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.7 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 1.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.9 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 10.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.9 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 4.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 5.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 1.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 57.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.7 | GO:0036019 | endolysosome(GO:0036019) endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.2 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.6 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.5 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 25.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.6 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 1.4 | 101.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 1.4 | 6.8 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.3 | 5.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 1.2 | 3.5 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.1 | 9.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.8 | 5.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.8 | 2.4 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.7 | 156.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.7 | 3.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.7 | 2.0 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.6 | 3.2 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.6 | 6.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.6 | 7.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.6 | 2.4 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.6 | 2.9 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.6 | 4.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.6 | 2.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.6 | 2.8 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.6 | 7.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.6 | 1.7 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.5 | 3.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.5 | 2.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.5 | 4.3 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.5 | 5.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.5 | 2.1 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.4 | 4.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.4 | 1.3 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.4 | 1.7 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.4 | 1.6 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.4 | 1.2 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.4 | 2.7 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.4 | 1.8 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.4 | 1.1 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.3 | 1.0 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.3 | 2.4 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.3 | 1.7 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.3 | 0.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.3 | 8.9 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 0.9 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.3 | 1.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.3 | 2.4 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 1.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 2.0 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.3 | 1.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.3 | 1.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.3 | 0.8 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.3 | 0.8 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.3 | 1.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 2.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 1.3 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.3 | 8.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 1.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.2 | 13.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 1.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.2 | 1.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 3.7 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 3.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.2 | 0.7 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.2 | 1.1 | GO:0004914 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 0.8 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.2 | 2.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 1.2 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 2.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 2.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 2.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 2.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 1.8 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 0.7 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 1.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 0.5 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.2 | 1.9 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 3.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 1.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 0.9 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.2 | 0.6 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.1 | 1.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 2.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.7 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 5.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.8 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.5 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 0.9 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.4 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.4 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.9 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.5 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 2.4 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.1 | 1.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.4 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 3.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 2.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.7 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 0.7 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.4 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.1 | 1.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 1.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 1.0 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 2.5 | GO:0001614 | G-protein coupled nucleotide receptor activity(GO:0001608) purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 2.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.3 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.3 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.1 | 1.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 2.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 2.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 2.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.6 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.5 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 1.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.3 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 1.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.6 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.9 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 10.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.4 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.2 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.1 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.4 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.4 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.6 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.1 | 1.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.9 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.5 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.7 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.5 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 1.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 4.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.3 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 1.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 4.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.4 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 1.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 1.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 1.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 2.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 7.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 4.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 2.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.7 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 1.0 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.9 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 4.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.7 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 6.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 2.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 3.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 1.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 4.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.4 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 1.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 4.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.9 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 2.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.8 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 18.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 1.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 2.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.2 | 4.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 10.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.3 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 2.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 6.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.7 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 2.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 0.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 7.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 1.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 9.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 7.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 3.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 16.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 9.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 2.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.7 | 10.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.4 | 14.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 9.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 5.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 20.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.3 | 9.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 5.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 2.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 4.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 7.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 3.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 15.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 6.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 1.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 2.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 10.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 1.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 1.4 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 2.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 2.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 3.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 2.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 10.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 3.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.8 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.0 | 1.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 2.7 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 2.0 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 1.1 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 3.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 4.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 2.2 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.4 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |