Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
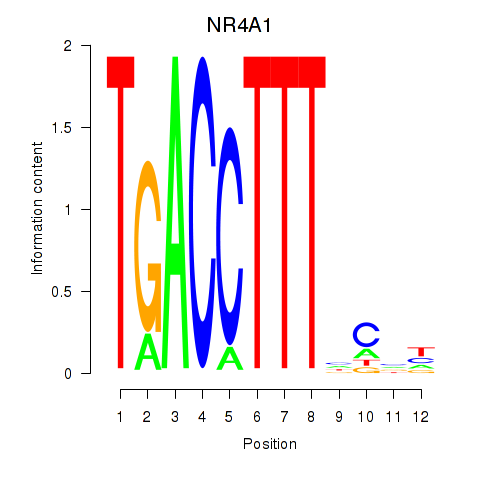
Results for NR4A1
Z-value: 0.82
Transcription factors associated with NR4A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A1
|
ENSG00000123358.20 | nuclear receptor subfamily 4 group A member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A1 | hg38_v1_chr12_+_52051402_52051459 | -0.16 | 3.8e-01 | Click! |
Activity profile of NR4A1 motif
Sorted Z-values of NR4A1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_4951080 | 4.68 |
ENST00000521811.5
ENST00000323997.10 ENST00000522249.5 ENST00000519584.5 ENST00000519602.6 |
ENO3
|
enolase 3 |
| chr17_+_4951758 | 3.73 |
ENST00000518175.1
|
ENO3
|
enolase 3 |
| chr11_+_116829898 | 2.31 |
ENST00000227667.8
ENST00000375345.3 |
APOC3
|
apolipoprotein C3 |
| chr12_+_109139397 | 2.20 |
ENST00000377854.9
ENST00000377848.7 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr19_+_4153616 | 1.72 |
ENST00000078445.7
ENST00000595923.5 ENST00000602257.5 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3 like 3 |
| chrX_+_2828921 | 1.66 |
ENST00000398806.8
|
GYG2
|
glycogenin 2 |
| chrX_+_2828808 | 1.66 |
ENST00000381163.7
|
GYG2
|
glycogenin 2 |
| chr1_+_236686454 | 1.60 |
ENST00000542672.6
ENST00000366578.6 ENST00000682015.1 ENST00000651275.1 |
ACTN2
|
actinin alpha 2 |
| chr4_-_110641920 | 1.27 |
ENST00000354925.6
ENST00000511990.1 ENST00000613094.4 ENST00000614423.4 ENST00000616641.4 ENST00000511837.5 |
PITX2
|
paired like homeodomain 2 |
| chr6_+_54083423 | 1.25 |
ENST00000460844.6
ENST00000370876.6 |
MLIP
|
muscular LMNA interacting protein |
| chr19_-_16496156 | 1.12 |
ENST00000269881.8
|
CALR3
|
calreticulin 3 |
| chr6_+_44158807 | 1.11 |
ENST00000532171.5
ENST00000398776.2 |
CAPN11
|
calpain 11 |
| chr3_+_46497970 | 1.10 |
ENST00000296142.4
|
RTP3
|
receptor transporter protein 3 |
| chr10_-_72954790 | 1.02 |
ENST00000373032.4
|
PLA2G12B
|
phospholipase A2 group XIIB |
| chr5_+_127703386 | 0.97 |
ENST00000514853.5
|
CCDC192
|
coiled-coil domain containing 192 |
| chr4_-_64409444 | 0.91 |
ENST00000381210.8
ENST00000507440.5 |
TECRL
|
trans-2,3-enoyl-CoA reductase like |
| chr12_+_21526287 | 0.88 |
ENST00000256969.7
|
SPX
|
spexin hormone |
| chr10_+_80413817 | 0.85 |
ENST00000372187.9
|
PRXL2A
|
peroxiredoxin like 2A |
| chr20_+_13008919 | 0.84 |
ENST00000399002.7
ENST00000434210.5 |
SPTLC3
|
serine palmitoyltransferase long chain base subunit 3 |
| chr7_-_107803215 | 0.83 |
ENST00000340010.10
ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 member 3 |
| chr4_-_88823306 | 0.83 |
ENST00000395002.6
|
FAM13A
|
family with sequence similarity 13 member A |
| chr4_+_99574812 | 0.81 |
ENST00000422897.6
ENST00000265517.10 |
MTTP
|
microsomal triglyceride transfer protein |
| chr6_+_108295037 | 0.81 |
ENST00000368977.9
ENST00000421954.5 |
AFG1L
|
AFG1 like ATPase |
| chr12_-_118052597 | 0.80 |
ENST00000535496.5
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr20_+_17699942 | 0.78 |
ENST00000427254.1
ENST00000377805.7 |
BANF2
|
BANF family member 2 |
| chr20_+_3071618 | 0.76 |
ENST00000217386.2
|
OXT
|
oxytocin/neurophysin I prepropeptide |
| chr16_-_30029429 | 0.76 |
ENST00000564806.1
|
TLCD3B
|
TLC domain containing 3B |
| chr20_+_13009246 | 0.76 |
ENST00000450297.1
|
SPTLC3
|
serine palmitoyltransferase long chain base subunit 3 |
| chr17_+_1742836 | 0.75 |
ENST00000324015.7
ENST00000450523.6 ENST00000453723.5 ENST00000453066.6 ENST00000382061.5 |
SERPINF2
|
serpin family F member 2 |
| chr20_+_17699960 | 0.75 |
ENST00000246090.6
|
BANF2
|
BANF family member 2 |
| chr12_+_57941499 | 0.74 |
ENST00000300145.4
|
ATP23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr5_-_127073467 | 0.74 |
ENST00000607731.5
ENST00000509733.7 ENST00000296662.10 ENST00000535381.6 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr4_-_40630826 | 0.72 |
ENST00000505414.5
ENST00000511598.5 |
RBM47
|
RNA binding motif protein 47 |
| chr1_+_40247926 | 0.69 |
ENST00000372766.4
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr14_+_37595847 | 0.65 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr11_+_87037820 | 0.65 |
ENST00000340353.11
|
TMEM135
|
transmembrane protein 135 |
| chr4_-_40630739 | 0.64 |
ENST00000511902.5
ENST00000505220.1 |
RBM47
|
RNA binding motif protein 47 |
| chr12_+_6535278 | 0.64 |
ENST00000396858.5
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr10_-_126670686 | 0.63 |
ENST00000488181.3
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr10_+_99782628 | 0.61 |
ENST00000648689.1
ENST00000647814.1 |
ABCC2
|
ATP binding cassette subfamily C member 2 |
| chr15_-_42491105 | 0.61 |
ENST00000565380.5
ENST00000564754.7 |
ZNF106
|
zinc finger protein 106 |
| chr4_-_88823214 | 0.59 |
ENST00000513837.5
ENST00000503556.5 |
FAM13A
|
family with sequence similarity 13 member A |
| chr7_-_38631356 | 0.59 |
ENST00000356264.7
ENST00000325590.9 |
AMPH
|
amphiphysin |
| chr10_+_12068945 | 0.57 |
ENST00000263035.9
|
DHTKD1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr1_+_228208024 | 0.57 |
ENST00000570156.7
ENST00000680850.1 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr1_+_228208054 | 0.57 |
ENST00000284548.16
ENST00000422127.5 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr12_-_16606795 | 0.56 |
ENST00000447609.5
|
LMO3
|
LIM domain only 3 |
| chr19_-_16496096 | 0.54 |
ENST00000600762.1
|
CALR3
|
calreticulin 3 |
| chr4_-_88823165 | 0.54 |
ENST00000508369.5
|
FAM13A
|
family with sequence similarity 13 member A |
| chr5_+_73813518 | 0.53 |
ENST00000296799.8
|
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr7_+_6009222 | 0.53 |
ENST00000400479.6
ENST00000223029.8 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex interacting multifunctional protein 2 |
| chr22_+_24495242 | 0.53 |
ENST00000382760.2
ENST00000326010.10 |
UPB1
|
beta-ureidopropionase 1 |
| chr19_+_6372757 | 0.52 |
ENST00000245812.8
|
ALKBH7
|
alkB homolog 7 |
| chr4_-_40630588 | 0.52 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr9_-_72953047 | 0.52 |
ENST00000297785.8
ENST00000376939.5 |
ALDH1A1
|
aldehyde dehydrogenase 1 family member A1 |
| chr4_-_73223082 | 0.51 |
ENST00000509867.6
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr6_+_133240587 | 0.51 |
ENST00000452339.6
|
EYA4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr3_-_48898813 | 0.50 |
ENST00000319017.5
ENST00000430379.5 |
SLC25A20
|
solute carrier family 25 member 20 |
| chr1_+_15736251 | 0.49 |
ENST00000294454.6
|
SLC25A34
|
solute carrier family 25 member 34 |
| chr11_+_87037915 | 0.49 |
ENST00000526733.5
ENST00000305494.6 ENST00000532959.5 |
TMEM135
|
transmembrane protein 135 |
| chr2_-_206159509 | 0.47 |
ENST00000423725.5
|
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr4_-_64409381 | 0.47 |
ENST00000509536.1
|
TECRL
|
trans-2,3-enoyl-CoA reductase like |
| chr11_+_66857056 | 0.46 |
ENST00000309602.5
ENST00000393952.3 |
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr10_+_12068964 | 0.45 |
ENST00000437298.1
|
DHTKD1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr2_-_206159410 | 0.44 |
ENST00000457011.5
ENST00000440274.5 ENST00000432169.5 ENST00000233190.11 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr3_-_139020926 | 0.43 |
ENST00000329447.5
|
PRR23B
|
proline rich 23B |
| chr12_-_52926459 | 0.42 |
ENST00000552150.5
|
KRT8
|
keratin 8 |
| chrX_-_54798253 | 0.41 |
ENST00000218436.7
|
ITIH6
|
inter-alpha-trypsin inhibitor heavy chain family member 6 |
| chr6_+_31927683 | 0.41 |
ENST00000456570.5
|
ENSG00000244255.5
|
novel complement component 2 (C2) and complement factor B (CFB) protein |
| chr20_+_58891981 | 0.40 |
ENST00000488652.6
ENST00000476935.6 ENST00000492907.6 ENST00000603546.2 |
GNAS
|
GNAS complex locus |
| chr5_-_143404536 | 0.37 |
ENST00000503201.1
ENST00000502892.5 |
NR3C1
|
nuclear receptor subfamily 3 group C member 1 |
| chr3_+_184319677 | 0.37 |
ENST00000441154.5
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma 1 |
| chr3_+_184320283 | 0.37 |
ENST00000428387.5
ENST00000434061.6 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma 1 |
| chr13_-_30306997 | 0.37 |
ENST00000380617.7
ENST00000441394.1 |
KATNAL1
|
katanin catalytic subunit A1 like 1 |
| chr6_-_116060859 | 0.37 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr2_+_174334947 | 0.37 |
ENST00000394967.3
|
SP9
|
Sp9 transcription factor |
| chrX_-_129523436 | 0.36 |
ENST00000371123.5
ENST00000371121.5 ENST00000371122.8 |
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr2_+_233307806 | 0.35 |
ENST00000447536.5
ENST00000409110.6 |
SAG
|
S-antigen visual arrestin |
| chr18_-_5197241 | 0.35 |
ENST00000434239.4
|
AKAIN1
|
A-kinase anchor inhibitor 1 |
| chr13_-_30306837 | 0.34 |
ENST00000414289.5
|
KATNAL1
|
katanin catalytic subunit A1 like 1 |
| chr2_-_43995999 | 0.33 |
ENST00000683213.1
ENST00000409946.6 |
LRPPRC
|
leucine rich pentatricopeptide repeat containing |
| chr12_+_18242955 | 0.33 |
ENST00000676171.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr5_-_74866914 | 0.33 |
ENST00000513277.5
|
FAM169A
|
family with sequence similarity 169 member A |
| chr12_-_16606102 | 0.33 |
ENST00000537304.6
|
LMO3
|
LIM domain only 3 |
| chr11_-_111923722 | 0.33 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B |
| chr14_+_35121814 | 0.32 |
ENST00000603611.5
|
PRORP
|
protein only RNase P catalytic subunit |
| chr12_-_16605939 | 0.31 |
ENST00000541295.5
ENST00000535535.5 |
LMO3
|
LIM domain only 3 |
| chr4_+_70242583 | 0.31 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr11_-_46616828 | 0.30 |
ENST00000529192.1
|
HARBI1
|
harbinger transposase derived 1 |
| chr12_+_32502178 | 0.30 |
ENST00000546442.5
ENST00000683182.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr17_-_68291116 | 0.30 |
ENST00000327268.8
ENST00000580666.6 |
SLC16A6
|
solute carrier family 16 member 6 |
| chr2_-_43995950 | 0.30 |
ENST00000683590.1
ENST00000683623.1 ENST00000682779.1 ENST00000682546.1 ENST00000683220.1 ENST00000683125.1 ENST00000683833.1 ENST00000683989.1 ENST00000682480.1 ENST00000260665.12 ENST00000682308.1 ENST00000682885.1 ENST00000409659.6 ENST00000447246.2 |
LRPPRC
|
leucine rich pentatricopeptide repeat containing |
| chr15_+_36594868 | 0.29 |
ENST00000566807.5
ENST00000643612.1 ENST00000567389.5 ENST00000562877.5 |
CDIN1
|
CDAN1 interacting nuclease 1 |
| chr19_+_17305801 | 0.29 |
ENST00000602206.1
ENST00000252602.2 |
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr14_-_95157270 | 0.28 |
ENST00000674628.1
ENST00000529720.1 ENST00000393063.6 ENST00000532939.3 |
DICER1
|
dicer 1, ribonuclease III |
| chr2_-_74440484 | 0.28 |
ENST00000305557.9
ENST00000233330.6 |
RTKN
|
rhotekin |
| chr7_+_131285041 | 0.28 |
ENST00000429546.5
|
MKLN1
|
muskelin 1 |
| chr5_-_124748800 | 0.28 |
ENST00000509799.5
|
ZNF608
|
zinc finger protein 608 |
| chr20_-_44187093 | 0.28 |
ENST00000342272.3
|
JPH2
|
junctophilin 2 |
| chr10_+_100969492 | 0.28 |
ENST00000519649.5
ENST00000518124.5 |
SEMA4G
|
semaphorin 4G |
| chr6_-_130215629 | 0.28 |
ENST00000532763.5
|
SAMD3
|
sterile alpha motif domain containing 3 |
| chr14_+_35122303 | 0.27 |
ENST00000604073.1
|
PRORP
|
protein only RNase P catalytic subunit |
| chr22_-_50578417 | 0.27 |
ENST00000312108.12
ENST00000395650.6 |
CPT1B
|
carnitine palmitoyltransferase 1B |
| chr11_+_3856182 | 0.26 |
ENST00000527651.5
|
STIM1
|
stromal interaction molecule 1 |
| chr8_+_143991454 | 0.26 |
ENST00000534791.5
ENST00000533044.5 |
GRINA
|
glutamate ionotropic receptor NMDA type subunit associated protein 1 |
| chr17_+_7219857 | 0.26 |
ENST00000583312.5
ENST00000350303.9 ENST00000584103.5 ENST00000356839.10 ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase very long chain |
| chr18_+_58196736 | 0.26 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr6_+_31927703 | 0.25 |
ENST00000418949.6
ENST00000299367.10 ENST00000383177.7 ENST00000477310.1 |
C2
ENSG00000244255.5
|
complement C2 novel complement component 2 (C2) and complement factor B (CFB) protein |
| chr18_+_13338599 | 0.24 |
ENST00000676672.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr3_+_113747022 | 0.24 |
ENST00000273398.8
ENST00000496747.5 ENST00000475322.1 |
ATP6V1A
|
ATPase H+ transporting V1 subunit A |
| chr19_+_13023958 | 0.24 |
ENST00000587760.5
ENST00000585575.5 |
NFIX
|
nuclear factor I X |
| chr6_+_31927510 | 0.23 |
ENST00000447952.6
|
C2
|
complement C2 |
| chr18_-_5197502 | 0.23 |
ENST00000580650.1
|
AKAIN1
|
A-kinase anchor inhibitor 1 |
| chr5_-_88268801 | 0.23 |
ENST00000506536.5
ENST00000512429.5 ENST00000514135.5 ENST00000296595.11 ENST00000509387.5 |
TMEM161B
|
transmembrane protein 161B |
| chr9_+_36572854 | 0.23 |
ENST00000543751.5
ENST00000536860.5 ENST00000541717.4 ENST00000536329.5 ENST00000536987.5 ENST00000298048.7 ENST00000495529.5 ENST00000545008.5 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr11_+_3855629 | 0.23 |
ENST00000526596.2
ENST00000300737.8 ENST00000616714.4 |
STIM1
|
stromal interaction molecule 1 |
| chr2_+_177392734 | 0.22 |
ENST00000680770.1
ENST00000637633.2 ENST00000679459.1 ENST00000409888.1 ENST00000264167.11 ENST00000642466.2 |
AGPS
|
alkylglycerone phosphate synthase |
| chr14_-_35122473 | 0.20 |
ENST00000557278.1
|
PPP2R3C
|
protein phosphatase 2 regulatory subunit B''gamma |
| chr16_-_81198766 | 0.20 |
ENST00000526632.5
|
PKD1L2
|
polycystin 1 like 2 (gene/pseudogene) |
| chr3_-_115147237 | 0.20 |
ENST00000357258.8
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr4_+_145680146 | 0.20 |
ENST00000438731.7
|
C4orf51
|
chromosome 4 open reading frame 51 |
| chr10_-_73591330 | 0.20 |
ENST00000451492.5
ENST00000681793.1 ENST00000680396.1 ENST00000413442.5 |
USP54
|
ubiquitin specific peptidase 54 |
| chr16_+_53207981 | 0.19 |
ENST00000565803.2
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr7_-_105876575 | 0.19 |
ENST00000318724.8
ENST00000419735.8 |
ATXN7L1
|
ataxin 7 like 1 |
| chr5_-_143405323 | 0.19 |
ENST00000514699.1
|
NR3C1
|
nuclear receptor subfamily 3 group C member 1 |
| chr18_+_46174055 | 0.19 |
ENST00000615553.1
|
C18orf25
|
chromosome 18 open reading frame 25 |
| chr18_+_46174014 | 0.18 |
ENST00000619301.4
ENST00000615052.5 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr16_+_55655960 | 0.18 |
ENST00000568943.6
|
SLC6A2
|
solute carrier family 6 member 2 |
| chr11_+_87037956 | 0.18 |
ENST00000525018.5
|
TMEM135
|
transmembrane protein 135 |
| chr15_+_75043263 | 0.18 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr17_-_38343810 | 0.17 |
ENST00000621958.1
ENST00000616987.5 |
GPR179
|
G protein-coupled receptor 179 |
| chrX_-_109625161 | 0.17 |
ENST00000372101.3
|
KCNE5
|
potassium voltage-gated channel subfamily E regulatory subunit 5 |
| chr1_+_176463164 | 0.17 |
ENST00000367661.7
ENST00000367662.5 |
PAPPA2
|
pappalysin 2 |
| chr12_+_32502114 | 0.16 |
ENST00000682739.1
ENST00000427716.7 ENST00000583694.2 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr6_-_43627302 | 0.16 |
ENST00000307114.11
|
GTPBP2
|
GTP binding protein 2 |
| chr16_-_31150058 | 0.16 |
ENST00000569305.1
ENST00000268281.9 ENST00000418068.6 |
PRSS36
|
serine protease 36 |
| chr1_-_197067234 | 0.15 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr17_-_7404039 | 0.15 |
ENST00000576017.1
ENST00000302422.4 |
TMEM256
|
transmembrane protein 256 |
| chr3_-_115147277 | 0.15 |
ENST00000675478.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr3_-_113746218 | 0.15 |
ENST00000497255.1
ENST00000240922.8 ENST00000478020.1 ENST00000493900.5 |
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr19_+_14028148 | 0.14 |
ENST00000431365.3
ENST00000585987.1 |
RLN3
|
relaxin 3 |
| chr11_+_72105924 | 0.14 |
ENST00000541899.2
|
ENSG00000284844.1
|
novel protein, ortholog of mouse Tomt |
| chr12_+_53985065 | 0.14 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr14_-_35122228 | 0.14 |
ENST00000555644.5
|
PPP2R3C
|
protein phosphatase 2 regulatory subunit B''gamma |
| chr6_+_69233023 | 0.13 |
ENST00000604969.1
ENST00000603207.1 |
ADGRB3
|
adhesion G protein-coupled receptor B3 |
| chr1_+_61404076 | 0.12 |
ENST00000357977.5
|
NFIA
|
nuclear factor I A |
| chr4_-_40630123 | 0.12 |
ENST00000514782.5
|
RBM47
|
RNA binding motif protein 47 |
| chr14_+_21098819 | 0.12 |
ENST00000418511.6
ENST00000554329.6 |
TMEM253
|
transmembrane protein 253 |
| chr3_+_14947568 | 0.11 |
ENST00000413118.5
ENST00000425241.5 |
NR2C2
|
nuclear receptor subfamily 2 group C member 2 |
| chr2_+_27059266 | 0.11 |
ENST00000441931.1
|
AGBL5
|
ATP/GTP binding protein like 5 |
| chr3_+_14947680 | 0.11 |
ENST00000435454.5
ENST00000323373.10 |
NR2C2
|
nuclear receptor subfamily 2 group C member 2 |
| chr19_+_49210734 | 0.11 |
ENST00000597316.1
|
TRPM4
|
transient receptor potential cation channel subfamily M member 4 |
| chr19_-_11529116 | 0.10 |
ENST00000592312.5
ENST00000590480.1 ENST00000585318.5 ENST00000270517.12 ENST00000252440.11 ENST00000417981.6 |
ECSIT
|
ECSIT signaling integrator |
| chr4_-_86358487 | 0.10 |
ENST00000641066.1
ENST00000512689.6 ENST00000641803.1 ENST00000640970.1 ENST00000641983.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr7_-_105876477 | 0.10 |
ENST00000478915.1
|
ATXN7L1
|
ataxin 7 like 1 |
| chr2_+_26346086 | 0.09 |
ENST00000613142.4
ENST00000260585.12 ENST00000447170.1 |
SELENOI
|
selenoprotein I |
| chr3_-_113746059 | 0.09 |
ENST00000477813.5
|
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr14_+_35122537 | 0.08 |
ENST00000604948.5
ENST00000321130.14 ENST00000534898.9 ENST00000605201.1 ENST00000250377.11 |
PRORP
|
protein only RNase P catalytic subunit |
| chr3_-_185821092 | 0.08 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr1_+_69567906 | 0.07 |
ENST00000651989.2
|
LRRC7
|
leucine rich repeat containing 7 |
| chr5_-_74866779 | 0.07 |
ENST00000510496.5
|
FAM169A
|
family with sequence similarity 169 member A |
| chr17_-_68291168 | 0.06 |
ENST00000582867.1
|
SLC16A6
|
solute carrier family 16 member 6 |
| chr17_-_40772162 | 0.06 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr15_-_42272825 | 0.06 |
ENST00000566474.5
|
TMEM87A
|
transmembrane protein 87A |
| chr1_-_161132577 | 0.06 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr6_+_31927486 | 0.05 |
ENST00000442278.6
|
C2
|
complement C2 |
| chr14_-_35121950 | 0.05 |
ENST00000554361.5
ENST00000261475.10 |
PPP2R3C
|
protein phosphatase 2 regulatory subunit B''gamma |
| chr3_+_138621207 | 0.05 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr7_-_126533850 | 0.04 |
ENST00000444921.3
|
GRM8
|
glutamate metabotropic receptor 8 |
| chr3_+_29280945 | 0.04 |
ENST00000434693.6
|
RBMS3
|
RNA binding motif single stranded interacting protein 3 |
| chr7_-_6009072 | 0.04 |
ENST00000642456.1
ENST00000642292.1 ENST00000441476.6 |
PMS2
|
PMS1 homolog 2, mismatch repair system component |
| chr7_-_81770428 | 0.04 |
ENST00000421558.1
|
HGF
|
hepatocyte growth factor |
| chr19_+_54415427 | 0.04 |
ENST00000301194.8
ENST00000376530.8 ENST00000445095.5 ENST00000376531.3 |
TTYH1
|
tweety family member 1 |
| chr20_-_45857196 | 0.03 |
ENST00000457981.5
ENST00000426915.1 ENST00000217455.9 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr3_+_138621225 | 0.02 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr4_-_56387422 | 0.02 |
ENST00000513376.5
ENST00000205214.11 ENST00000502617.1 ENST00000451613.5 ENST00000602986.5 |
AASDH
|
aminoadipate-semialdehyde dehydrogenase |
| chr8_-_91040814 | 0.02 |
ENST00000520014.1
ENST00000285419.8 |
PIP4P2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
| chr12_+_50401108 | 0.02 |
ENST00000517559.5
ENST00000552445.1 |
LARP4
|
La ribonucleoprotein 4 |
| chr7_+_12570712 | 0.02 |
ENST00000417018.1
ENST00000297029.10 |
SCIN
|
scinderin |
| chr15_-_40828699 | 0.02 |
ENST00000299174.10
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1 regulatory inhibitor subunit 14D |
| chr11_-_113706330 | 0.02 |
ENST00000540540.5
ENST00000538955.5 ENST00000545579.6 |
TMPRSS5
|
transmembrane serine protease 5 |
| chr3_-_139044892 | 0.01 |
ENST00000413199.1
|
PRR23C
|
proline rich 23C |
| chr19_-_40750302 | 0.01 |
ENST00000598485.6
ENST00000378313.7 ENST00000470681.5 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr3_-_157503574 | 0.01 |
ENST00000494677.5
ENST00000468233.5 |
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr3_-_113746185 | 0.01 |
ENST00000616174.1
|
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr3_+_101724633 | 0.00 |
ENST00000494050.5
|
CEP97
|
centrosomal protein 97 |
| chr6_+_63636481 | 0.00 |
ENST00000509330.5
|
PHF3
|
PHD finger protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:2000910 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.5 | 1.6 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.4 | 1.3 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.3 | 2.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.3 | 0.8 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.2 | 0.6 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.2 | 0.5 | GO:0019483 | beta-alanine biosynthetic process(GO:0019483) |
| 0.2 | 2.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.5 | GO:0006844 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.2 | 0.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 0.6 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.1 | 0.5 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 8.4 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.9 | GO:1904306 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.1 | 1.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.8 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.5 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 1.6 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 1.1 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.1 | 0.5 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.5 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.8 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.2 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 3.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 0.8 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.7 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.5 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 1.7 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.0 | 0.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.0 | 0.3 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.4 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 1.3 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.3 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.4 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0036404 | conversion of ds siRNA to ss siRNA involved in RNA interference(GO:0033168) conversion of ds siRNA to ss siRNA(GO:0036404) |
| 0.0 | 0.4 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 1.0 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 1.0 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0090494 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 2.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.9 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 0.7 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 1.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0033167 | ARC complex(GO:0033167) |
| 0.0 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 2.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.7 | 3.3 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.5 | 2.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.3 | 2.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 1.6 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 0.6 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.2 | 1.6 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.5 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 1.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 1.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.4 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.2 | GO:0052858 | peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) |
| 0.1 | 0.7 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.6 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.5 | GO:0018479 | androgen binding(GO:0005497) benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 1.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.0 | 1.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 1.0 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.4 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 1.4 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 3.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 2.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |