Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
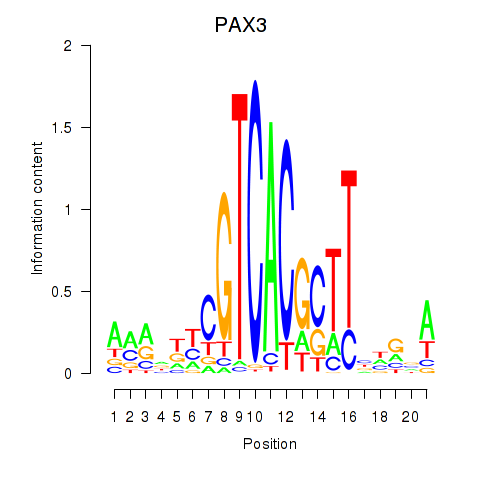
Results for PAX3
Z-value: 1.11
Transcription factors associated with PAX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX3
|
ENSG00000135903.20 | paired box 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX3 | hg38_v1_chr2_-_222298808_222298895 | 0.36 | 4.5e-02 | Click! |
Activity profile of PAX3 motif
Sorted Z-values of PAX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_71144818 | 2.23 |
ENST00000536169.5
ENST00000358741.4 ENST00000395855.6 ENST00000374051.7 |
NLGN3
|
neuroligin 3 |
| chr2_-_216860042 | 2.12 |
ENST00000236979.2
|
TNP1
|
transition protein 1 |
| chr22_-_43862480 | 2.10 |
ENST00000330884.9
|
SULT4A1
|
sulfotransferase family 4A member 1 |
| chr11_+_73647549 | 1.99 |
ENST00000227214.10
ENST00000398494.8 ENST00000543085.5 |
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr12_+_6951345 | 1.96 |
ENST00000318974.14
ENST00000541698.5 ENST00000542462.1 |
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chr12_+_6951271 | 1.87 |
ENST00000456013.5
|
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chr12_+_6951250 | 1.76 |
ENST00000538715.5
|
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chr4_-_167234579 | 1.73 |
ENST00000502330.5
ENST00000357154.7 ENST00000421836.6 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr5_+_127703386 | 1.67 |
ENST00000514853.5
|
CCDC192
|
coiled-coil domain containing 192 |
| chr4_-_167234426 | 1.63 |
ENST00000541354.5
ENST00000509854.5 ENST00000512681.5 ENST00000357545.9 ENST00000510741.5 ENST00000510403.5 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr15_+_51681483 | 1.62 |
ENST00000542355.6
ENST00000220478.8 ENST00000558709.1 |
SCG3
|
secretogranin III |
| chr22_+_41976760 | 1.62 |
ENST00000396426.7
ENST00000406029.5 |
SEPTIN3
|
septin 3 |
| chr15_+_37934626 | 1.61 |
ENST00000559502.5
ENST00000558148.5 ENST00000319669.5 ENST00000558158.5 |
TMCO5A
|
transmembrane and coiled-coil domains 5A |
| chr22_+_18110802 | 1.61 |
ENST00000330423.8
|
TUBA8
|
tubulin alpha 8 |
| chr11_+_73647645 | 1.56 |
ENST00000545798.5
ENST00000539157.5 ENST00000546251.5 ENST00000535582.5 ENST00000538227.5 |
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr11_+_65354745 | 1.54 |
ENST00000309880.6
|
TIGD3
|
tigger transposable element derived 3 |
| chrX_+_37990773 | 1.48 |
ENST00000341016.5
|
H2AP
|
H2A.P histone |
| chr10_-_124744254 | 1.42 |
ENST00000280780.6
|
FAM53B
|
family with sequence similarity 53 member B |
| chr1_+_145719483 | 1.42 |
ENST00000369288.7
|
CD160
|
CD160 molecule |
| chr7_-_142962206 | 1.40 |
ENST00000460479.2
ENST00000476829.5 ENST00000355265.7 |
KEL
|
Kell metallo-endopeptidase (Kell blood group) |
| chr8_-_30912998 | 1.38 |
ENST00000643185.2
|
TEX15
|
testis expressed 15, meiosis and synapsis associated |
| chr2_-_157327699 | 1.30 |
ENST00000397283.6
|
ERMN
|
ermin |
| chr1_+_6034980 | 1.27 |
ENST00000378092.6
ENST00000472700.7 |
KCNAB2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr17_-_75154503 | 1.24 |
ENST00000409753.8
ENST00000581874.1 |
JPT1
|
Jupiter microtubule associated homolog 1 |
| chr3_+_14675128 | 1.15 |
ENST00000435614.5
ENST00000253697.8 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr1_+_84164370 | 1.12 |
ENST00000446538.5
ENST00000610703.4 ENST00000370682.7 ENST00000394838.6 ENST00000432111.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chrY_+_18546691 | 1.11 |
ENST00000309834.8
ENST00000307393.3 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor Y-linked 1 |
| chr5_+_174046659 | 1.11 |
ENST00000519717.1
|
NSG2
|
neuronal vesicle trafficking associated 2 |
| chr7_+_100219236 | 1.07 |
ENST00000317271.2
|
PVRIG
|
PVR related immunoglobulin domain containing |
| chr12_-_10409757 | 1.07 |
ENST00000309384.2
|
KLRC4
|
killer cell lectin like receptor C4 |
| chr11_+_66083971 | 1.06 |
ENST00000533756.5
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr15_-_82952683 | 1.05 |
ENST00000450735.7
ENST00000304231.12 |
HOMER2
|
homer scaffold protein 2 |
| chr19_-_37594746 | 1.05 |
ENST00000593133.5
ENST00000590751.5 ENST00000358744.3 ENST00000328550.6 ENST00000451802.7 |
ZNF571
|
zinc finger protein 571 |
| chr1_+_84164293 | 1.04 |
ENST00000370684.5
ENST00000436133.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_+_167868948 | 1.03 |
ENST00000392690.3
|
B3GALT1
|
beta-1,3-galactosyltransferase 1 |
| chr10_-_124744280 | 1.02 |
ENST00000337318.8
|
FAM53B
|
family with sequence similarity 53 member B |
| chr12_-_75209814 | 1.00 |
ENST00000549446.6
|
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr4_-_167234266 | 0.99 |
ENST00000511269.5
ENST00000506697.5 ENST00000512042.1 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr11_+_73647701 | 0.98 |
ENST00000543524.5
|
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr1_-_205211535 | 0.98 |
ENST00000367161.7
|
DSTYK
|
dual serine/threonine and tyrosine protein kinase |
| chr1_-_205211669 | 0.98 |
ENST00000367162.8
|
DSTYK
|
dual serine/threonine and tyrosine protein kinase |
| chr14_+_21749163 | 0.97 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr7_+_107660819 | 0.97 |
ENST00000644269.2
|
SLC26A4
|
solute carrier family 26 member 4 |
| chr20_-_17531366 | 0.96 |
ENST00000377873.8
|
BFSP1
|
beaded filament structural protein 1 |
| chr19_+_48019726 | 0.94 |
ENST00000593413.1
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr4_-_167234552 | 0.94 |
ENST00000512648.5
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr12_+_10010627 | 0.93 |
ENST00000338896.11
ENST00000396502.5 |
CLEC12B
|
C-type lectin domain family 12 member B |
| chr15_-_43330537 | 0.91 |
ENST00000305641.7
ENST00000567039.1 |
LCMT2
|
leucine carboxyl methyltransferase 2 |
| chr14_-_20590823 | 0.91 |
ENST00000556526.1
|
RNASE12
|
ribonuclease A family member 12 (inactive) |
| chr9_-_89178810 | 0.89 |
ENST00000375835.9
|
SHC3
|
SHC adaptor protein 3 |
| chr5_-_157652432 | 0.89 |
ENST00000265007.11
|
SOX30
|
SRY-box transcription factor 30 |
| chr11_+_112167366 | 0.86 |
ENST00000530752.5
ENST00000280358.5 |
TEX12
|
testis expressed 12 |
| chr7_+_142492121 | 0.86 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6 |
| chr8_+_116938180 | 0.86 |
ENST00000378279.4
|
AARD
|
alanine and arginine rich domain containing protein |
| chr18_+_61333424 | 0.84 |
ENST00000262717.9
|
CDH20
|
cadherin 20 |
| chr13_-_60013178 | 0.81 |
ENST00000498416.2
ENST00000465066.5 |
DIAPH3
|
diaphanous related formin 3 |
| chr15_-_52191387 | 0.80 |
ENST00000261837.12
|
GNB5
|
G protein subunit beta 5 |
| chr4_-_170003738 | 0.80 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibril associated protein 3 like |
| chr9_-_35650902 | 0.80 |
ENST00000259608.8
ENST00000618781.1 |
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr1_+_99264275 | 0.80 |
ENST00000457765.6
|
PLPPR4
|
phospholipid phosphatase related 4 |
| chr15_+_43330646 | 0.80 |
ENST00000562188.7
ENST00000428046.7 ENST00000389651.8 |
ADAL
|
adenosine deaminase like |
| chr9_-_112175185 | 0.79 |
ENST00000355396.7
|
SUSD1
|
sushi domain containing 1 |
| chr18_+_26226417 | 0.78 |
ENST00000269142.10
|
TAF4B
|
TATA-box binding protein associated factor 4b |
| chr15_-_52569126 | 0.76 |
ENST00000569723.5
ENST00000567669.5 ENST00000569281.6 ENST00000563566.5 ENST00000567830.1 |
ARPP19
|
cAMP regulated phosphoprotein 19 |
| chr1_+_145719468 | 0.76 |
ENST00000616463.1
|
CD160
|
CD160 molecule |
| chr12_+_10307950 | 0.76 |
ENST00000543420.5
ENST00000543777.5 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr12_-_53200443 | 0.75 |
ENST00000550743.6
|
ITGB7
|
integrin subunit beta 7 |
| chr19_+_15010720 | 0.74 |
ENST00000292574.4
|
CCDC105
|
coiled-coil domain containing 105 |
| chr7_+_142384328 | 0.74 |
ENST00000390361.3
|
TRBV7-3
|
T cell receptor beta variable 7-3 |
| chr5_+_172194172 | 0.73 |
ENST00000398186.5
|
EFCAB9
|
EF-hand calcium binding domain 9 |
| chr17_-_3557798 | 0.73 |
ENST00000301365.8
ENST00000572519.1 ENST00000576742.6 |
TRPV3
|
transient receptor potential cation channel subfamily V member 3 |
| chr5_+_148268741 | 0.72 |
ENST00000398450.5
|
SPINK13
|
serine peptidase inhibitor Kazal type 13 |
| chr7_-_151633182 | 0.70 |
ENST00000476632.2
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr12_+_10307818 | 0.70 |
ENST00000350274.9
ENST00000336164.9 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr5_-_181261078 | 0.70 |
ENST00000611618.1
|
TRIM52
|
tripartite motif containing 52 |
| chr18_+_26226472 | 0.67 |
ENST00000578121.5
|
TAF4B
|
TATA-box binding protein associated factor 4b |
| chr11_+_7597182 | 0.65 |
ENST00000528883.5
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr6_+_133241318 | 0.65 |
ENST00000430974.6
ENST00000355286.12 ENST00000355167.8 ENST00000431403.3 |
EYA4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr12_+_8989535 | 0.64 |
ENST00000356986.8
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr10_+_122980392 | 0.64 |
ENST00000406217.8
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr9_+_33795551 | 0.61 |
ENST00000379405.4
|
PRSS3
|
serine protease 3 |
| chr14_+_51847116 | 0.61 |
ENST00000553560.5
|
GNG2
|
G protein subunit gamma 2 |
| chrX_-_15854791 | 0.61 |
ENST00000545766.7
ENST00000380291.5 ENST00000672987.1 ENST00000329235.6 |
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chr1_+_206897435 | 0.61 |
ENST00000391929.7
ENST00000294984.7 ENST00000611909.4 ENST00000367093.3 |
IL24
|
interleukin 24 |
| chr7_+_142352802 | 0.61 |
ENST00000634605.1
|
TRBV7-2
|
T cell receptor beta variable 7-2 |
| chr19_+_37594867 | 0.61 |
ENST00000316433.9
ENST00000590588.5 ENST00000586134.5 ENST00000586792.5 |
ZNF540
|
zinc finger protein 540 |
| chr12_+_8989612 | 0.61 |
ENST00000266551.8
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr1_+_12464912 | 0.60 |
ENST00000543766.2
|
VPS13D
|
vacuolar protein sorting 13 homolog D |
| chr17_-_61591192 | 0.59 |
ENST00000521764.3
|
NACA2
|
nascent polypeptide associated complex subunit alpha 2 |
| chr14_+_44995455 | 0.59 |
ENST00000555874.5
|
TOGARAM1
|
TOG array regulator of axonemal microtubules 1 |
| chr19_-_35228699 | 0.59 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187 member B |
| chr1_-_183418364 | 0.59 |
ENST00000287713.7
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr5_+_50665899 | 0.58 |
ENST00000505697.6
|
PARP8
|
poly(ADP-ribose) polymerase family member 8 |
| chr4_-_48114523 | 0.58 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr14_+_22207502 | 0.58 |
ENST00000390461.2
|
TRAV34
|
T cell receptor alpha variable 34 |
| chrX_+_154542194 | 0.58 |
ENST00000618670.4
|
IKBKG
|
inhibitor of nuclear factor kappa B kinase regulatory subunit gamma |
| chr11_+_66291887 | 0.57 |
ENST00000327259.5
|
TMEM151A
|
transmembrane protein 151A |
| chr7_+_107661002 | 0.56 |
ENST00000440056.1
|
SLC26A4
|
solute carrier family 26 member 4 |
| chr3_-_119660580 | 0.56 |
ENST00000493094.6
ENST00000264231.7 ENST00000468801.1 |
POPDC2
|
popeye domain containing 2 |
| chr3_-_51779187 | 0.55 |
ENST00000398780.5
ENST00000668964.1 ENST00000667863.2 ENST00000647442.1 |
IQCF6
|
IQ motif containing F6 |
| chr17_+_37375974 | 0.55 |
ENST00000615133.2
ENST00000611038.4 |
C17orf78
|
chromosome 17 open reading frame 78 |
| chr19_-_20565769 | 0.55 |
ENST00000427401.9
|
ZNF737
|
zinc finger protein 737 |
| chr19_-_40090860 | 0.55 |
ENST00000599972.1
ENST00000450241.6 ENST00000595687.6 ENST00000340963.9 |
ZNF780A
|
zinc finger protein 780A |
| chr9_-_111759508 | 0.54 |
ENST00000394777.8
ENST00000394779.7 |
SHOC1
|
shortage in chiasmata 1 |
| chr2_-_182522556 | 0.53 |
ENST00000435564.5
|
PDE1A
|
phosphodiesterase 1A |
| chr2_-_218659468 | 0.53 |
ENST00000450560.1
ENST00000449707.5 ENST00000440934.2 |
ZNF142
|
zinc finger protein 142 |
| chr19_-_43883964 | 0.53 |
ENST00000587539.2
|
ZNF404
|
zinc finger protein 404 |
| chr7_-_108243234 | 0.53 |
ENST00000417701.5
|
NRCAM
|
neuronal cell adhesion molecule |
| chr17_-_62806632 | 0.52 |
ENST00000583803.1
ENST00000456609.6 |
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chrX_-_119565362 | 0.52 |
ENST00000320339.8
ENST00000536133.2 ENST00000644802.2 |
STEEP1
|
STING1 ER exit protein 1 |
| chr9_-_97922487 | 0.52 |
ENST00000455506.1
ENST00000375117.8 ENST00000375119.8 ENST00000611338.4 |
TRMO
|
tRNA methyltransferase O |
| chr1_+_50048014 | 0.52 |
ENST00000448907.7
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr9_+_128787243 | 0.50 |
ENST00000372648.10
|
TBC1D13
|
TBC1 domain family member 13 |
| chr2_-_44361754 | 0.49 |
ENST00000409272.5
ENST00000410081.5 ENST00000541738.5 |
PREPL
|
prolyl endopeptidase like |
| chr1_+_108746654 | 0.49 |
ENST00000370008.4
|
STXBP3
|
syntaxin binding protein 3 |
| chr1_-_101025763 | 0.49 |
ENST00000342173.11
ENST00000488176.1 ENST00000370109.8 |
DPH5
|
diphthamide biosynthesis 5 |
| chrX_+_30247139 | 0.49 |
ENST00000397548.4
|
MAGEB1
|
MAGE family member B1 |
| chr1_-_205121986 | 0.48 |
ENST00000367164.1
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr2_-_182242031 | 0.47 |
ENST00000358139.6
|
PDE1A
|
phosphodiesterase 1A |
| chr5_+_148268830 | 0.46 |
ENST00000511106.5
|
SPINK13
|
serine peptidase inhibitor Kazal type 13 |
| chr1_-_205121964 | 0.46 |
ENST00000264515.11
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr19_-_47545261 | 0.46 |
ENST00000595558.1
ENST00000263351.9 |
ZNF541
|
zinc finger protein 541 |
| chr1_-_51878711 | 0.45 |
ENST00000352171.12
|
NRDC
|
nardilysin convertase |
| chr19_-_45076465 | 0.45 |
ENST00000303809.7
|
ZNF296
|
zinc finger protein 296 |
| chr2_-_44361485 | 0.44 |
ENST00000438314.1
ENST00000409411.6 ENST00000409936.5 |
PREPL
|
prolyl endopeptidase like |
| chr10_+_116545907 | 0.44 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr2_-_68252482 | 0.43 |
ENST00000234310.8
|
PPP3R1
|
protein phosphatase 3 regulatory subunit B, alpha |
| chr2_-_218659592 | 0.42 |
ENST00000411696.7
|
ZNF142
|
zinc finger protein 142 |
| chr1_-_26890237 | 0.42 |
ENST00000431781.2
ENST00000374135.9 |
GPN2
|
GPN-loop GTPase 2 |
| chr3_-_52239082 | 0.42 |
ENST00000499914.2
ENST00000678838.1 ENST00000305533.10 ENST00000678330.1 |
TWF2
|
twinfilin actin binding protein 2 |
| chr14_+_51847145 | 0.42 |
ENST00000615906.4
|
GNG2
|
G protein subunit gamma 2 |
| chr6_+_35259470 | 0.42 |
ENST00000469195.5
|
ZNF76
|
zinc finger protein 76 |
| chrX_+_103628692 | 0.41 |
ENST00000372626.7
|
TCEAL1
|
transcription elongation factor A like 1 |
| chr2_-_44361555 | 0.41 |
ENST00000409957.5
|
PREPL
|
prolyl endopeptidase like |
| chr19_-_50333504 | 0.41 |
ENST00000474951.1
|
KCNC3
|
potassium voltage-gated channel subfamily C member 3 |
| chrX_+_103628959 | 0.40 |
ENST00000372625.8
ENST00000372624.3 |
TCEAL1
|
transcription elongation factor A like 1 |
| chr2_+_170178136 | 0.40 |
ENST00000409044.7
ENST00000408978.9 |
MYO3B
|
myosin IIIB |
| chr10_-_102419693 | 0.40 |
ENST00000611678.4
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr2_-_182522703 | 0.39 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A |
| chr9_+_101398841 | 0.38 |
ENST00000339664.7
ENST00000374861.7 ENST00000259395.4 ENST00000615466.1 |
ZNF189
|
zinc finger protein 189 |
| chr3_+_49689531 | 0.37 |
ENST00000432042.5
ENST00000454491.5 ENST00000327697.11 |
RNF123
|
ring finger protein 123 |
| chr16_-_71489276 | 0.37 |
ENST00000564230.5
ENST00000565637.5 ENST00000288177.10 |
ZNF19
|
zinc finger protein 19 |
| chr3_+_184300564 | 0.37 |
ENST00000435761.5
ENST00000439383.5 |
PSMD2
|
proteasome 26S subunit, non-ATPase 2 |
| chr19_-_51501755 | 0.36 |
ENST00000291707.8
|
SIGLEC12
|
sialic acid binding Ig like lectin 12 |
| chr17_-_41382298 | 0.36 |
ENST00000394001.3
|
KRT34
|
keratin 34 |
| chr13_+_97953652 | 0.35 |
ENST00000460070.6
ENST00000481455.6 ENST00000261574.10 ENST00000651721.2 ENST00000493281.6 ENST00000463157.6 ENST00000471898.5 ENST00000489058.6 ENST00000481689.6 |
IPO5
|
importin 5 |
| chr9_-_21217311 | 0.34 |
ENST00000380216.1
|
IFNA16
|
interferon alpha 16 |
| chr14_-_59465327 | 0.33 |
ENST00000395116.1
|
GPR135
|
G protein-coupled receptor 135 |
| chr19_-_36916268 | 0.33 |
ENST00000391711.8
|
ZNF829
|
zinc finger protein 829 |
| chr19_-_40090921 | 0.32 |
ENST00000595508.1
ENST00000414720.6 ENST00000455521.5 ENST00000595773.5 ENST00000683561.1 |
ENSG00000269749.1
ZNF780A
|
novel transcript zinc finger protein 780A |
| chr4_-_86357722 | 0.32 |
ENST00000641341.1
ENST00000642038.1 ENST00000641116.1 ENST00000641767.1 ENST00000639242.1 ENST00000638313.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr3_+_112990956 | 0.32 |
ENST00000383678.8
ENST00000383677.7 ENST00000619116.4 |
GTPBP8
|
GTP binding protein 8 (putative) |
| chr1_-_243186382 | 0.32 |
ENST00000522895.1
|
CEP170
|
centrosomal protein 170 |
| chr7_-_19773569 | 0.31 |
ENST00000422233.5
ENST00000433641.5 ENST00000405844.6 |
TMEM196
|
transmembrane protein 196 |
| chr2_-_96208815 | 0.31 |
ENST00000443962.1
ENST00000337288.10 |
STARD7
|
StAR related lipid transfer domain containing 7 |
| chr15_-_42491105 | 0.30 |
ENST00000565380.5
ENST00000564754.7 |
ZNF106
|
zinc finger protein 106 |
| chr1_-_51878799 | 0.30 |
ENST00000354831.11
ENST00000544028.5 |
NRDC
|
nardilysin convertase |
| chr8_+_103414703 | 0.29 |
ENST00000616836.4
ENST00000297579.9 |
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr10_+_102245371 | 0.29 |
ENST00000676513.1
ENST00000676939.1 ENST00000677947.1 ENST00000677247.1 ENST00000369983.4 ENST00000678351.1 ENST00000679238.1 ENST00000677439.1 ENST00000677240.1 ENST00000677618.1 ENST00000673650.1 ENST00000674034.1 ENST00000676993.1 |
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr2_+_73385811 | 0.28 |
ENST00000613296.6
ENST00000614410.4 |
ALMS1
|
ALMS1 centrosome and basal body associated protein |
| chr11_+_31369834 | 0.28 |
ENST00000465995.6
|
DNAJC24
|
DnaJ heat shock protein family (Hsp40) member C24 |
| chr1_-_13198130 | 0.28 |
ENST00000638454.1
|
PRAMEF13
|
PRAME family member 13 |
| chr9_-_101398590 | 0.27 |
ENST00000374865.5
|
MRPL50
|
mitochondrial ribosomal protein L50 |
| chr14_-_100375333 | 0.26 |
ENST00000557297.5
ENST00000555813.5 ENST00000392882.7 ENST00000557135.5 ENST00000556698.5 ENST00000554509.5 ENST00000555410.5 |
WARS1
|
tryptophanyl-tRNA synthetase 1 |
| chr16_+_29790715 | 0.26 |
ENST00000561482.5
ENST00000569636.6 ENST00000160827.9 |
KIF22
|
kinesin family member 22 |
| chr20_-_25585517 | 0.26 |
ENST00000422516.5
ENST00000278886.11 |
NINL
|
ninein like |
| chr17_-_40417873 | 0.26 |
ENST00000423485.6
|
TOP2A
|
DNA topoisomerase II alpha |
| chr12_+_112125531 | 0.26 |
ENST00000549358.5
ENST00000257604.9 ENST00000548092.5 ENST00000412615.7 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr9_+_128787331 | 0.25 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family member 13 |
| chr9_-_86282511 | 0.25 |
ENST00000375991.9
ENST00000326094.4 |
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr6_+_14117764 | 0.24 |
ENST00000379153.4
|
CD83
|
CD83 molecule |
| chr2_+_44361897 | 0.23 |
ENST00000378494.8
ENST00000402247.5 ENST00000407131.5 ENST00000403853.7 |
CAMKMT
|
calmodulin-lysine N-methyltransferase |
| chr10_+_89327989 | 0.23 |
ENST00000679923.1
ENST00000680085.1 ENST00000371818.9 ENST00000680779.1 |
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chr9_-_36401198 | 0.23 |
ENST00000377885.6
|
RNF38
|
ring finger protein 38 |
| chr1_+_154272589 | 0.22 |
ENST00000457918.6
ENST00000483970.6 ENST00000328703.12 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr11_-_73982830 | 0.21 |
ENST00000536983.5
ENST00000663595.2 ENST00000310473.9 |
UCP2
|
uncoupling protein 2 |
| chr7_-_148883474 | 0.21 |
ENST00000476773.5
|
EZH2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr7_+_129375830 | 0.20 |
ENST00000466993.5
|
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr10_+_89327977 | 0.20 |
ENST00000681277.1
|
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chr21_-_36079382 | 0.20 |
ENST00000399201.5
|
SETD4
|
SET domain containing 4 |
| chr6_+_127266875 | 0.19 |
ENST00000610162.5
ENST00000608991.5 ENST00000610153.1 |
RNF146
|
ring finger protein 146 |
| chr13_+_31739520 | 0.19 |
ENST00000298386.7
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr16_-_18375069 | 0.18 |
ENST00000545114.5
|
NPIPA9
|
nuclear pore complex interacting protein family, member A9 |
| chr6_+_108295037 | 0.18 |
ENST00000368977.9
ENST00000421954.5 |
AFG1L
|
AFG1 like ATPase |
| chr6_+_133241566 | 0.18 |
ENST00000531901.5
|
EYA4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr4_-_103198371 | 0.17 |
ENST00000611174.4
ENST00000380026.8 |
CENPE
|
centromere protein E |
| chr2_-_24972032 | 0.17 |
ENST00000534855.5
|
DNAJC27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr6_-_31972123 | 0.17 |
ENST00000337523.10
|
DXO
|
decapping exoribonuclease |
| chrX_-_49073989 | 0.17 |
ENST00000376386.3
ENST00000553851.3 |
PRAF2
|
PRA1 domain family member 2 |
| chr2_+_151357583 | 0.15 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6 |
| chr1_+_247507058 | 0.15 |
ENST00000527084.5
ENST00000536561.5 ENST00000527541.5 ENST00000366491.6 ENST00000366489.5 ENST00000526896.5 |
GCSAML
|
germinal center associated signaling and motility like |
| chr7_+_142469521 | 0.14 |
ENST00000390371.3
|
TRBV6-6
|
T cell receptor beta variable 6-6 |
| chr17_-_69150062 | 0.14 |
ENST00000522787.5
ENST00000521538.5 |
ABCA10
|
ATP binding cassette subfamily A member 10 |
| chr4_+_15374541 | 0.14 |
ENST00000382383.7
ENST00000429690.5 |
C1QTNF7
|
C1q and TNF related 7 |
| chr22_+_38705737 | 0.13 |
ENST00000484657.5
|
GTPBP1
|
GTP binding protein 1 |
| chr6_+_35259703 | 0.13 |
ENST00000373953.8
ENST00000339411.5 |
ZNF76
|
zinc finger protein 76 |
| chr11_+_123902167 | 0.13 |
ENST00000641687.1
|
OR8D4
|
olfactory receptor family 8 subfamily D member 4 |
| chr6_+_127266832 | 0.13 |
ENST00000356799.6
ENST00000368314.6 |
RNF146
|
ring finger protein 146 |
| chr19_+_37594830 | 0.13 |
ENST00000589117.5
|
ZNF540
|
zinc finger protein 540 |
| chr9_-_21240002 | 0.13 |
ENST00000380222.4
|
IFNA14
|
interferon alpha 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.6 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.6 | 2.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.4 | 2.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.3 | 0.9 | GO:0002769 | natural killer cell inhibitory signaling pathway(GO:0002769) |
| 0.2 | 1.0 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.2 | 5.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 2.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 0.9 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.2 | 1.4 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 1.2 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.2 | 0.8 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.6 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 2.0 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.6 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 1.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.8 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.4 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.7 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.1 | 1.0 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.6 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.3 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.8 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.2 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 1.1 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 1.0 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 1.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 2.1 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 4.8 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.8 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 2.4 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 0.4 | GO:0032532 | regulation of microvillus length(GO:0032532) regulation of cell projection size(GO:0032536) |
| 0.0 | 1.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.7 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 4.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.5 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 1.1 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 1.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 1.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 1.0 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.5 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.8 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.1 | GO:0030718 | inner cell mass cell fate commitment(GO:0001827) germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 1.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0050992 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 1.3 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.7 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.4 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 1.5 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.0 | 0.8 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.6 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.6 | GO:0043276 | anoikis(GO:0043276) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.3 | 2.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 1.3 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.2 | 0.8 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.9 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.4 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.6 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 1.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 2.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 1.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.1 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.3 | 1.5 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 1.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 5.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.6 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 2.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 5.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.6 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.1 | 1.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 2.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.5 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 2.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 2.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 1.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 1.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 1.0 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.9 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 1.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.6 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 2.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.3 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 2.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 2.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 1.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME MEIOSIS | Genes involved in Meiosis |