Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for PAX7_NOBOX
Z-value: 1.01
Transcription factors associated with PAX7_NOBOX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
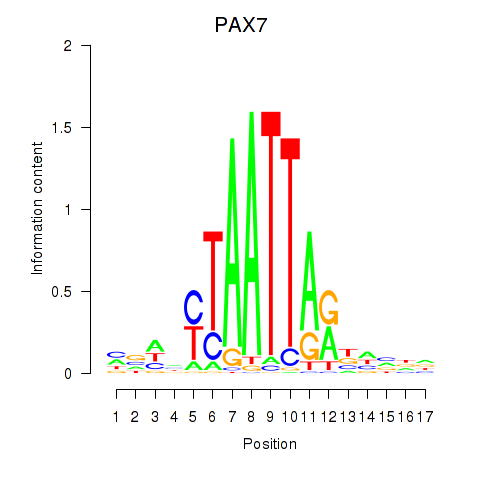
PAX7
|
ENSG00000009709.12 | paired box 7 |
|
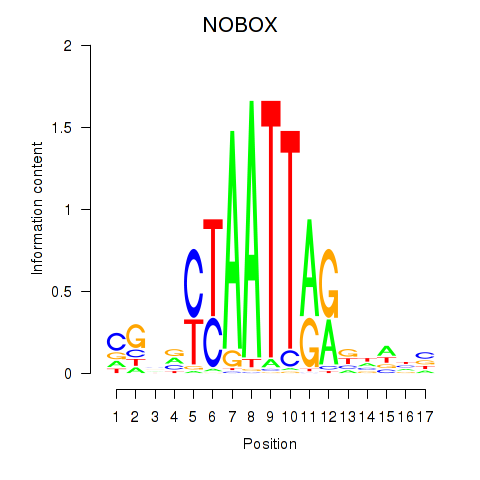
NOBOX
|
ENSG00000106410.15 | NOBOX oogenesis homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX7 | hg38_v1_chr1_+_18631513_18631529 | -0.35 | 5.0e-02 | Click! |
| NOBOX | hg38_v1_chr7_-_144403693_144403693 | 0.12 | 5.0e-01 | Click! |
Activity profile of PAX7_NOBOX motif
Sorted Z-values of PAX7_NOBOX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX7_NOBOX
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.6 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 1.2 | 7.3 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 1.1 | 3.4 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.9 | 2.6 | GO:0046333 | octopamine biosynthetic process(GO:0006589) homoiothermy(GO:0042309) octopamine metabolic process(GO:0046333) |
| 0.8 | 2.5 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.8 | 2.3 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.7 | 5.0 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.7 | 2.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.6 | 3.7 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.6 | 1.7 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.5 | 1.8 | GO:0052363 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.5 | 1.4 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.4 | 1.3 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.4 | 2.2 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.4 | 1.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.4 | 2.5 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.4 | 1.3 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.3 | 3.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.3 | 0.9 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) |
| 0.3 | 5.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.3 | 0.8 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.3 | 1.1 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.3 | 1.3 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.3 | 4.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 0.7 | GO:0002665 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.2 | 1.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.2 | 0.7 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.2 | 0.9 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.2 | 1.6 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.2 | 0.9 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.2 | 0.6 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 1.4 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.2 | 0.6 | GO:0036290 | protein trans-autophosphorylation(GO:0036290) peptidyl-serine trans-autophosphorylation(GO:1990579) |
| 0.2 | 0.6 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.2 | 1.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.2 | 1.6 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.2 | 0.3 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.2 | 0.3 | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) |
| 0.2 | 0.9 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.6 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 2.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.7 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.6 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.1 | 1.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 5.4 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.4 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.1 | 2.0 | GO:0032074 | regulation of endodeoxyribonuclease activity(GO:0032071) negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 0.5 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.5 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 1.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.8 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 2.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.2 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 0.3 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.6 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.6 | GO:1990418 | response to sodium phosphate(GO:1904383) response to insulin-like growth factor stimulus(GO:1990418) |
| 0.1 | 0.3 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.4 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 2.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 1.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.8 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.9 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 0.8 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 3.0 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.6 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.4 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.3 | GO:0060922 | cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.1 | 0.7 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.9 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 1.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.5 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.1 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 0.2 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.5 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 1.0 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 0.4 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.4 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.4 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 2.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.1 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.0 | 1.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.7 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 1.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 8.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) regulation of mitotic cell cycle DNA replication(GO:1903463) negative regulation of mitotic cell cycle DNA replication(GO:1903464) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 2.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.8 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 1.5 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 1.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.6 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 2.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.3 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.5 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.6 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 2.5 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.2 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.0 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 1.7 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 0.8 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 1.2 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 3.6 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 1.2 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.7 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.9 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.3 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 1.0 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.4 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 1.0 | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle(GO:0007091) |
| 0.0 | 0.7 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.3 | GO:0001780 | neutrophil homeostasis(GO:0001780) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 3.3 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.4 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.4 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.2 | GO:2000580 | nuclear migration along microfilament(GO:0031022) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.0 | 0.7 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 0.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.8 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.3 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.9 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.0 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) adaptation of signaling pathway(GO:0023058) light adaption(GO:0036367) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.4 | 1.4 | GO:0097196 | Shu complex(GO:0097196) |
| 0.3 | 0.9 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.2 | 1.7 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 1.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 0.6 | GO:1990332 | Ire1 complex(GO:1990332) |
| 0.2 | 1.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 1.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 1.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 3.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 2.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.4 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 0.3 | GO:0033150 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.1 | 4.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.6 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 5.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.2 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.1 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.9 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.1 | 0.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 7.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.9 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 4.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.0 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 4.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 6.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 1.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0030892 | mitotic cohesin complex(GO:0030892) |
| 0.0 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 7.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 4.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.8 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.6 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.9 | 2.6 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.6 | 5.0 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.6 | 3.7 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.6 | 2.2 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.5 | 1.6 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.5 | 1.6 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.5 | 2.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 1.3 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.4 | 1.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.4 | 1.6 | GO:0008513 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.3 | 0.9 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.3 | 0.9 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.3 | 1.8 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 8.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 1.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 0.8 | GO:0033265 | choline binding(GO:0033265) |
| 0.2 | 1.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 0.8 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.2 | 0.6 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.2 | 0.9 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.2 | 1.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 0.9 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.2 | 0.5 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 0.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 3.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 3.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.3 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 1.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.7 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.9 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 3.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 1.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 1.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.9 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.3 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 2.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 2.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 1.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 1.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.3 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 2.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.2 | GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity(GO:0003860) |
| 0.1 | 1.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 2.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 2.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.3 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 4.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.0 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 12.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.6 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 2.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 1.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 1.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.3 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 2.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0000252 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) sterol-4-alpha-carboxylate 3-dehydrogenase (decarboxylating) activity(GO:0047012) |
| 0.0 | 0.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.0 | 1.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.4 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 2.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0070975 | FHA domain binding(GO:0070975) |
| 0.0 | 5.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0030290 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.0 | 0.0 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 2.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 2.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 4.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 3.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 7.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 4.0 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 4.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 3.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 7.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 5.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 1.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 0.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 2.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 4.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 3.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 3.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 0.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 2.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 1.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.8 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 2.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 5.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |