Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for PAX8
Z-value: 0.78
Transcription factors associated with PAX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX8
|
ENSG00000125618.18 | paired box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX8 | hg38_v1_chr2_-_113241683_113241697 | -0.06 | 7.6e-01 | Click! |
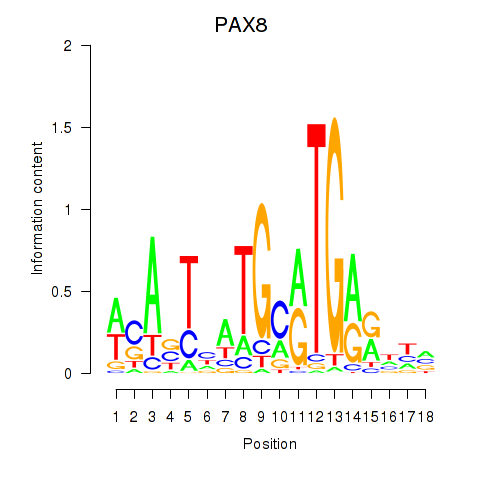
Activity profile of PAX8 motif
Sorted Z-values of PAX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51750798 | 2.06 |
ENST00000600815.1
|
FPR1
|
formyl peptide receptor 1 |
| chr10_+_133527355 | 1.70 |
ENST00000252945.8
ENST00000421586.5 ENST00000418356.1 |
CYP2E1
|
cytochrome P450 family 2 subfamily E member 1 |
| chr1_-_27626229 | 1.59 |
ENST00000399173.5
|
FGR
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr1_-_27626496 | 1.56 |
ENST00000374003.7
|
FGR
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr1_-_27626106 | 1.54 |
ENST00000457296.5
|
FGR
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr1_-_161631152 | 1.45 |
ENST00000421702.3
ENST00000650385.1 |
FCGR3B
|
Fc fragment of IgG receptor IIIb |
| chr7_+_142352802 | 1.42 |
ENST00000634605.1
|
TRBV7-2
|
T cell receptor beta variable 7-2 |
| chr9_-_114074969 | 1.42 |
ENST00000466610.6
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr3_+_186717348 | 1.37 |
ENST00000447445.1
ENST00000287611.8 ENST00000644859.2 |
KNG1
|
kininogen 1 |
| chr6_+_160121809 | 1.35 |
ENST00000366963.9
|
SLC22A1
|
solute carrier family 22 member 1 |
| chr1_-_161631032 | 1.31 |
ENST00000534776.1
ENST00000613418.4 ENST00000614870.4 |
FCGR3B
|
Fc fragment of IgG receptor IIIb |
| chr11_+_60455839 | 1.26 |
ENST00000532491.5
ENST00000532073.5 ENST00000345732.9 ENST00000534668.6 ENST00000528313.1 ENST00000533306.6 ENST00000674194.1 |
MS4A1
|
membrane spanning 4-domains A1 |
| chr6_+_160121859 | 1.23 |
ENST00000324965.8
ENST00000457470.6 |
SLC22A1
|
solute carrier family 22 member 1 |
| chr19_-_7702124 | 1.22 |
ENST00000597921.6
|
FCER2
|
Fc fragment of IgE receptor II |
| chr6_-_44265541 | 1.17 |
ENST00000619360.6
|
NFKBIE
|
NFKB inhibitor epsilon |
| chr10_+_69088096 | 1.13 |
ENST00000242465.4
|
SRGN
|
serglycin |
| chr7_+_142592928 | 1.11 |
ENST00000616518.1
|
TRBV15
|
T cell receptor beta variable 15 |
| chr14_-_22819721 | 1.08 |
ENST00000554517.5
ENST00000285850.11 ENST00000397529.6 ENST00000555702.5 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr12_-_95996302 | 1.08 |
ENST00000261208.8
ENST00000538703.5 ENST00000541929.5 |
HAL
|
histidine ammonia-lyase |
| chr19_-_7702139 | 0.94 |
ENST00000346664.9
|
FCER2
|
Fc fragment of IgE receptor II |
| chr7_+_142391884 | 0.94 |
ENST00000390363.2
|
TRBV9
|
T cell receptor beta variable 9 |
| chr22_-_50085414 | 0.94 |
ENST00000311597.10
|
MLC1
|
modulator of VRAC current 1 |
| chr12_-_10420550 | 0.93 |
ENST00000381903.2
ENST00000396439.7 |
KLRC3
|
killer cell lectin like receptor C3 |
| chr7_+_142720652 | 0.92 |
ENST00000390400.2
|
TRBV28
|
T cell receptor beta variable 28 |
| chr14_+_94619313 | 0.88 |
ENST00000621603.1
|
SERPINA3
|
serpin family A member 3 |
| chr6_-_24935942 | 0.87 |
ENST00000645100.1
ENST00000643898.2 ENST00000613507.4 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr22_-_50085331 | 0.86 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr6_+_24126186 | 0.85 |
ENST00000378478.5
ENST00000378491.9 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr16_-_3256587 | 0.84 |
ENST00000536379.5
ENST00000541159.5 ENST00000339854.8 ENST00000219596.6 |
MEFV
|
MEFV innate immuity regulator, pyrin |
| chr6_+_160702238 | 0.81 |
ENST00000366924.6
ENST00000308192.14 ENST00000418964.1 |
PLG
|
plasminogen |
| chr14_+_22462932 | 0.80 |
ENST00000390477.2
|
TRDC
|
T cell receptor delta constant |
| chr12_+_124993633 | 0.79 |
ENST00000341446.9
ENST00000671775.2 |
BRI3BP
|
BRI3 binding protein |
| chr19_+_35448251 | 0.78 |
ENST00000599180.3
|
FFAR2
|
free fatty acid receptor 2 |
| chr10_+_12349685 | 0.78 |
ENST00000378845.5
|
CAMK1D
|
calcium/calmodulin dependent protein kinase ID |
| chr1_+_203765168 | 0.77 |
ENST00000367217.5
ENST00000442561.7 |
LAX1
|
lymphocyte transmembrane adaptor 1 |
| chr22_-_50085378 | 0.76 |
ENST00000442311.1
|
MLC1
|
modulator of VRAC current 1 |
| chr7_-_14974773 | 0.75 |
ENST00000437998.1
|
DGKB
|
diacylglycerol kinase beta |
| chr14_+_58331253 | 0.74 |
ENST00000417477.2
|
ARID4A
|
AT-rich interaction domain 4A |
| chr17_+_7558296 | 0.73 |
ENST00000438470.5
ENST00000436057.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr14_+_21978440 | 0.73 |
ENST00000390443.3
|
TRAV8-6
|
T cell receptor alpha variable 8-6 |
| chr19_+_41796616 | 0.72 |
ENST00000344550.4
|
CEACAM3
|
CEA cell adhesion molecule 3 |
| chr4_+_36281591 | 0.72 |
ENST00000639862.2
ENST00000357504.7 |
DTHD1
|
death domain containing 1 |
| chr6_+_41228339 | 0.71 |
ENST00000448827.6
ENST00000341495.7 |
TREML4
|
triggering receptor expressed on myeloid cells like 4 |
| chr12_+_75480745 | 0.70 |
ENST00000266659.8
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr2_+_17539964 | 0.70 |
ENST00000457525.5
|
VSNL1
|
visinin like 1 |
| chr14_-_91244669 | 0.69 |
ENST00000650645.1
|
GPR68
|
G protein-coupled receptor 68 |
| chr16_-_3577288 | 0.68 |
ENST00000324659.12
|
NLRC3
|
NLR family CARD domain containing 3 |
| chr7_-_144835981 | 0.67 |
ENST00000360057.7
ENST00000378099.7 ENST00000639328.1 |
TPK1
|
thiamin pyrophosphokinase 1 |
| chr17_+_7558712 | 0.66 |
ENST00000338784.9
ENST00000625791.2 |
TNFSF13
|
TNF superfamily member 13 |
| chr8_+_119067239 | 0.66 |
ENST00000332843.3
|
COLEC10
|
collectin subfamily member 10 |
| chr4_+_118034480 | 0.65 |
ENST00000296499.6
|
NDST3
|
N-deacetylase and N-sulfotransferase 3 |
| chr6_-_32816910 | 0.65 |
ENST00000447394.1
ENST00000438763.7 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr9_-_89405926 | 0.65 |
ENST00000433650.5
|
SEMA4D
|
semaphorin 4D |
| chr12_+_57434778 | 0.65 |
ENST00000309668.3
|
INHBC
|
inhibin subunit beta C |
| chr11_+_60971777 | 0.64 |
ENST00000542157.5
ENST00000433107.6 ENST00000352009.9 ENST00000452451.6 |
CD6
|
CD6 molecule |
| chr17_+_7558774 | 0.62 |
ENST00000396545.4
|
TNFSF13
|
TNF superfamily member 13 |
| chr14_-_91244508 | 0.62 |
ENST00000535815.5
ENST00000529102.1 |
GPR68
|
G protein-coupled receptor 68 |
| chrX_-_153876327 | 0.62 |
ENST00000458029.1
|
L1CAM
|
L1 cell adhesion molecule |
| chr1_+_158930778 | 0.62 |
ENST00000458222.5
|
PYHIN1
|
pyrin and HIN domain family member 1 |
| chr8_-_52409743 | 0.62 |
ENST00000276480.11
|
ST18
|
ST18 C2H2C-type zinc finger transcription factor |
| chr12_+_75480800 | 0.61 |
ENST00000456650.7
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr19_+_51225059 | 0.61 |
ENST00000436584.6
ENST00000421133.6 ENST00000262262.5 ENST00000391796.7 |
CD33
|
CD33 molecule |
| chr12_-_57479848 | 0.59 |
ENST00000393791.8
ENST00000552249.1 |
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr13_-_25172278 | 0.59 |
ENST00000515384.2
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr16_-_3577375 | 0.59 |
ENST00000359128.10
|
NLRC3
|
NLR family CARD domain containing 3 |
| chr1_-_84690735 | 0.59 |
ENST00000422026.1
|
SSX2IP
|
SSX family member 2 interacting protein |
| chr6_-_134318097 | 0.59 |
ENST00000367858.10
ENST00000533224.1 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr14_+_20781139 | 0.58 |
ENST00000304677.3
|
RNASE6
|
ribonuclease A family member k6 |
| chr12_-_110445540 | 0.58 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex subunit 3 |
| chr12_-_10435940 | 0.58 |
ENST00000381901.5
ENST00000381902.7 ENST00000539033.1 |
KLRC2
ENSG00000255641.1
|
killer cell lectin like receptor C2 novel protein |
| chr22_-_43862480 | 0.57 |
ENST00000330884.9
|
SULT4A1
|
sulfotransferase family 4A member 1 |
| chr13_+_30735523 | 0.57 |
ENST00000380490.5
|
ALOX5AP
|
arachidonate 5-lipoxygenase activating protein |
| chr22_+_20774092 | 0.57 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chr3_+_190388120 | 0.57 |
ENST00000456423.2
ENST00000264734.3 |
CLDN16
|
claudin 16 |
| chr4_-_69787955 | 0.55 |
ENST00000512870.1
|
SULT1B1
|
sulfotransferase family 1B member 1 |
| chr2_+_190469486 | 0.54 |
ENST00000444317.1
|
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr1_+_116372647 | 0.54 |
ENST00000418797.5
|
ATP1A1
|
ATPase Na+/K+ transporting subunit alpha 1 |
| chr19_-_15934410 | 0.54 |
ENST00000326742.12
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11 |
| chr15_+_89088417 | 0.51 |
ENST00000569550.5
ENST00000565066.5 ENST00000565973.5 ENST00000352732.10 |
ABHD2
|
abhydrolase domain containing 2, acylglycerol lipase |
| chr17_+_7558258 | 0.51 |
ENST00000483039.5
ENST00000380535.8 ENST00000396542.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr6_+_167122742 | 0.51 |
ENST00000341935.9
ENST00000349984.6 |
CCR6
|
C-C motif chemokine receptor 6 |
| chr1_+_160739239 | 0.50 |
ENST00000368043.8
|
SLAMF7
|
SLAM family member 7 |
| chr2_-_174847525 | 0.49 |
ENST00000295497.12
ENST00000652036.1 ENST00000444394.6 ENST00000650731.1 |
CHN1
|
chimerin 1 |
| chr7_+_95485898 | 0.49 |
ENST00000428113.5
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr11_-_58575846 | 0.48 |
ENST00000395074.7
|
LPXN
|
leupaxin |
| chr22_-_42070778 | 0.47 |
ENST00000396398.8
ENST00000403363.5 ENST00000402937.1 |
NAGA
|
alpha-N-acetylgalactosaminidase |
| chr7_+_150514851 | 0.47 |
ENST00000313543.5
|
GIMAP7
|
GTPase, IMAP family member 7 |
| chr8_-_52409787 | 0.47 |
ENST00000517580.5
|
ST18
|
ST18 C2H2C-type zinc finger transcription factor |
| chr17_-_63931354 | 0.46 |
ENST00000647774.1
|
ENSG00000285947.1
|
novel protein |
| chr19_+_41797147 | 0.45 |
ENST00000596544.1
|
CEACAM3
|
CEA cell adhesion molecule 3 |
| chr17_+_7558465 | 0.43 |
ENST00000349228.8
|
TNFSF13
|
TNF superfamily member 13 |
| chr2_-_2326210 | 0.43 |
ENST00000647755.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr8_+_58411333 | 0.43 |
ENST00000399598.7
|
UBXN2B
|
UBX domain protein 2B |
| chr6_+_43298326 | 0.43 |
ENST00000372574.7
|
SLC22A7
|
solute carrier family 22 member 7 |
| chr2_-_2326161 | 0.42 |
ENST00000649810.1
ENST00000648318.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr14_-_106335613 | 0.42 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr20_-_46684467 | 0.42 |
ENST00000372121.5
|
SLC13A3
|
solute carrier family 13 member 3 |
| chr1_-_198540674 | 0.41 |
ENST00000489986.1
ENST00000367382.6 |
ATP6V1G3
|
ATPase H+ transporting V1 subunit G3 |
| chr4_+_68815991 | 0.40 |
ENST00000265403.12
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase family 2 member B10 |
| chr1_+_89524819 | 0.40 |
ENST00000439853.6
ENST00000330947.7 ENST00000449440.5 ENST00000640258.1 |
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr19_+_35351552 | 0.40 |
ENST00000246553.3
|
FFAR1
|
free fatty acid receptor 1 |
| chr12_-_57016517 | 0.40 |
ENST00000441881.5
ENST00000458521.7 |
TAC3
|
tachykinin precursor 3 |
| chr2_-_174847765 | 0.39 |
ENST00000443238.6
|
CHN1
|
chimerin 1 |
| chr14_-_106771020 | 0.39 |
ENST00000617374.2
|
IGHV2-70
|
immunoglobulin heavy variable 2-70 |
| chr12_-_128823942 | 0.38 |
ENST00000266771.10
|
SLC15A4
|
solute carrier family 15 member 4 |
| chr18_-_27990256 | 0.38 |
ENST00000675173.1
|
CDH2
|
cadherin 2 |
| chr7_+_150322639 | 0.37 |
ENST00000343855.6
|
ZBED6CL
|
ZBED6 C-terminal like |
| chr2_+_87748087 | 0.37 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2 |
| chr18_+_45825666 | 0.37 |
ENST00000389474.8
|
SIGLEC15
|
sialic acid binding Ig like lectin 15 |
| chr1_+_89524871 | 0.36 |
ENST00000639264.1
|
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr4_+_2798564 | 0.36 |
ENST00000504294.5
|
SH3BP2
|
SH3 domain binding protein 2 |
| chr5_+_140868945 | 0.35 |
ENST00000398640.7
|
PCDHA11
|
protocadherin alpha 11 |
| chr16_-_28506826 | 0.35 |
ENST00000356897.1
|
IL27
|
interleukin 27 |
| chr15_+_23565666 | 0.34 |
ENST00000314520.6
ENST00000679144.1 ENST00000564592.2 ENST00000677119.1 ENST00000647595.1 |
MKRN3
|
makorin ring finger protein 3 |
| chr1_-_186680411 | 0.34 |
ENST00000367468.10
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 |
| chr2_-_2326378 | 0.34 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr9_-_96383675 | 0.33 |
ENST00000375257.2
ENST00000375259.9 ENST00000253270.13 |
SLC35D2
|
solute carrier family 35 member D2 |
| chr1_-_173207322 | 0.33 |
ENST00000281834.4
|
TNFSF4
|
TNF superfamily member 4 |
| chr1_-_184037695 | 0.33 |
ENST00000361927.9
ENST00000649786.1 |
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr2_-_37147734 | 0.32 |
ENST00000405334.5
|
EIF2AK2
|
eukaryotic translation initiation factor 2 alpha kinase 2 |
| chr11_+_123358416 | 0.32 |
ENST00000638157.1
|
GRAMD1B
|
GRAM domain containing 1B |
| chr1_+_100352451 | 0.32 |
ENST00000361544.11
ENST00000370124.8 ENST00000336454.5 |
CDC14A
|
cell division cycle 14A |
| chr20_+_59996335 | 0.32 |
ENST00000244049.7
ENST00000350849.10 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr9_-_95506601 | 0.31 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr7_-_99679987 | 0.31 |
ENST00000222982.8
ENST00000439761.3 ENST00000339843.6 |
CYP3A5
|
cytochrome P450 family 3 subfamily A member 5 |
| chr11_+_17260353 | 0.31 |
ENST00000530527.5
|
NUCB2
|
nucleobindin 2 |
| chr18_-_60372767 | 0.31 |
ENST00000299766.5
|
MC4R
|
melanocortin 4 receptor |
| chr9_-_95507199 | 0.30 |
ENST00000553011.5
ENST00000551845.5 |
PTCH1
|
patched 1 |
| chr13_+_24270841 | 0.30 |
ENST00000454083.1
|
SPATA13
|
spermatogenesis associated 13 |
| chr19_+_18007182 | 0.30 |
ENST00000595712.6
|
ARRDC2
|
arrestin domain containing 2 |
| chr1_-_169764648 | 0.30 |
ENST00000454271.1
ENST00000609271.1 |
ENSG00000230704.1
SELE
|
novel transcript selectin E |
| chr2_+_79025678 | 0.29 |
ENST00000393897.6
|
REG3G
|
regenerating family member 3 gamma |
| chr5_+_90640718 | 0.29 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr11_+_63974578 | 0.28 |
ENST00000314133.4
ENST00000535431.1 |
COX8A
ENSG00000256100.1
|
cytochrome c oxidase subunit 8A novel transcript |
| chr16_-_67453693 | 0.28 |
ENST00000564615.5
|
ATP6V0D1
|
ATPase H+ transporting V0 subunit d1 |
| chr2_+_79025696 | 0.28 |
ENST00000272324.10
|
REG3G
|
regenerating family member 3 gamma |
| chr21_-_7789509 | 0.28 |
ENST00000646133.1
|
SMIM34B
|
small integral membrane protein 34B |
| chrX_-_102155790 | 0.28 |
ENST00000543160.5
ENST00000333643.4 |
BEX5
|
brain expressed X-linked 5 |
| chr3_-_161372821 | 0.27 |
ENST00000617024.1
ENST00000359175.8 |
SPTSSB
|
serine palmitoyltransferase small subunit B |
| chr1_+_40450053 | 0.27 |
ENST00000484445.5
ENST00000411995.6 |
ZFP69B
|
ZFP69 zinc finger protein B |
| chr9_-_21228222 | 0.27 |
ENST00000413767.2
|
IFNA17
|
interferon alpha 17 |
| chr20_+_4171709 | 0.27 |
ENST00000379460.6
|
SMOX
|
spermine oxidase |
| chr19_+_41350911 | 0.27 |
ENST00000539627.5
|
TMEM91
|
transmembrane protein 91 |
| chr10_+_102152380 | 0.27 |
ENST00000605788.6
ENST00000488254.6 ENST00000461421.5 ENST00000476468.5 ENST00000370007.5 |
NOLC1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr9_-_136996555 | 0.26 |
ENST00000494426.2
|
CLIC3
|
chloride intracellular channel 3 |
| chr22_+_20116099 | 0.26 |
ENST00000430524.6
|
RANBP1
|
RAN binding protein 1 |
| chr2_+_119544420 | 0.26 |
ENST00000413369.8
|
CFAP221
|
cilia and flagella associated protein 221 |
| chrX_-_153875847 | 0.26 |
ENST00000361699.8
ENST00000361981.7 |
L1CAM
|
L1 cell adhesion molecule |
| chr15_-_29452799 | 0.25 |
ENST00000560082.1
|
FAM189A1
|
family with sequence similarity 189 member A1 |
| chr17_-_75270409 | 0.25 |
ENST00000618645.5
|
MIF4GD
|
MIF4G domain containing |
| chr17_-_75270999 | 0.25 |
ENST00000579194.6
ENST00000580717.5 ENST00000577542.5 ENST00000579612.5 ENST00000245551.9 ENST00000578305.5 |
MIF4GD
|
MIF4G domain containing |
| chr1_+_207034366 | 0.25 |
ENST00000545806.5
ENST00000618513.4 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr21_+_43867215 | 0.24 |
ENST00000448287.5
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr14_+_21653835 | 0.24 |
ENST00000641524.1
|
OR4E2
|
olfactory receptor family 4 subfamily E member 2 |
| chr5_-_88877967 | 0.23 |
ENST00000508610.5
ENST00000636294.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr12_-_56741535 | 0.23 |
ENST00000647707.1
|
ENSG00000285625.1
|
novel protein |
| chr4_+_74365136 | 0.23 |
ENST00000244869.3
|
EREG
|
epiregulin |
| chrX_-_153875979 | 0.23 |
ENST00000407935.6
ENST00000439496.5 |
L1CAM
|
L1 cell adhesion molecule |
| chr5_+_55851349 | 0.23 |
ENST00000652347.2
|
IL31RA
|
interleukin 31 receptor A |
| chr11_-_129192198 | 0.23 |
ENST00000310343.13
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr2_-_24328113 | 0.23 |
ENST00000622089.4
|
ITSN2
|
intersectin 2 |
| chr1_-_248574757 | 0.22 |
ENST00000328782.3
|
OR2T34
|
olfactory receptor family 2 subfamily T member 34 |
| chr13_-_102759059 | 0.22 |
ENST00000322527.4
|
CCDC168
|
coiled-coil domain containing 168 |
| chr1_-_58546693 | 0.22 |
ENST00000456980.5
ENST00000482274.2 ENST00000453710.1 ENST00000371226.8 ENST00000419242.5 ENST00000426139.5 |
OMA1
|
OMA1 zinc metallopeptidase |
| chr17_-_75270710 | 0.22 |
ENST00000581777.2
|
MIF4GD
|
MIF4G domain containing |
| chr17_-_75271223 | 0.22 |
ENST00000579297.5
ENST00000580571.5 ENST00000325102.13 |
MIF4GD
|
MIF4G domain containing |
| chr2_+_79025709 | 0.22 |
ENST00000409471.1
|
REG3G
|
regenerating family member 3 gamma |
| chr6_+_72212802 | 0.22 |
ENST00000401910.7
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr8_-_133256830 | 0.21 |
ENST00000674925.1
|
NDRG1
|
N-myc downstream regulated 1 |
| chr10_+_102152169 | 0.21 |
ENST00000405356.5
|
NOLC1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr4_+_37977269 | 0.21 |
ENST00000446803.6
|
TBC1D1
|
TBC1 domain family member 1 |
| chr12_-_54258275 | 0.21 |
ENST00000552562.1
|
CBX5
|
chromobox 5 |
| chr5_+_140855882 | 0.21 |
ENST00000562220.2
ENST00000307360.6 ENST00000506939.6 |
PCDHA10
|
protocadherin alpha 10 |
| chr6_+_72212887 | 0.21 |
ENST00000523963.5
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr11_-_129192291 | 0.20 |
ENST00000682385.1
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr6_+_29306626 | 0.19 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr2_+_172860038 | 0.19 |
ENST00000538974.5
ENST00000540783.5 |
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chrX_-_132094263 | 0.19 |
ENST00000370879.5
|
FRMD7
|
FERM domain containing 7 |
| chr2_-_127858107 | 0.19 |
ENST00000409955.1
ENST00000272645.9 |
POLR2D
|
RNA polymerase II subunit D |
| chr12_-_10169155 | 0.19 |
ENST00000539518.5
|
OLR1
|
oxidized low density lipoprotein receptor 1 |
| chr10_-_43397220 | 0.19 |
ENST00000477108.5
ENST00000544000.5 |
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chrX_-_152992195 | 0.19 |
ENST00000361887.5
ENST00000452693.5 ENST00000439251.3 |
PNMA5
|
PNMA family member 5 |
| chr19_-_1513577 | 0.18 |
ENST00000585804.2
|
ADAMTSL5
|
ADAMTS like 5 |
| chr18_-_55401026 | 0.18 |
ENST00000562607.5
|
TCF4
|
transcription factor 4 |
| chr9_+_136979042 | 0.18 |
ENST00000446677.2
|
PTGDS
|
prostaglandin D2 synthase |
| chr9_-_132079856 | 0.18 |
ENST00000651555.1
ENST00000651950.1 ENST00000357028.6 ENST00000474263.1 ENST00000292035.10 |
MED27
|
mediator complex subunit 27 |
| chr1_-_74512611 | 0.18 |
ENST00000294635.5
|
LRRC53
|
leucine rich repeat containing 53 |
| chrX_-_101052054 | 0.18 |
ENST00000372939.5
ENST00000372935.5 |
TRMT2B
|
tRNA methyltransferase 2 homolog B |
| chr2_+_90209873 | 0.18 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr13_-_46020487 | 0.17 |
ENST00000464597.2
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr21_-_34511243 | 0.17 |
ENST00000399284.1
|
KCNE1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr14_-_106154113 | 0.17 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr2_+_42169332 | 0.17 |
ENST00000402711.6
ENST00000318522.10 |
EML4
|
EMAP like 4 |
| chr1_-_8526025 | 0.17 |
ENST00000464972.5
|
RERE
|
arginine-glutamic acid dipeptide repeats |
| chr2_+_170180125 | 0.17 |
ENST00000484338.6
|
MYO3B
|
myosin IIIB |
| chr17_-_75271205 | 0.17 |
ENST00000649805.1
|
MIF4GD
|
MIF4G domain containing |
| chr4_+_71339014 | 0.17 |
ENST00000340595.4
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr4_+_71339037 | 0.16 |
ENST00000512686.5
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr5_-_135399863 | 0.16 |
ENST00000510038.1
ENST00000304332.8 |
MACROH2A1
|
macroH2A.1 histone |
| chr9_+_122614738 | 0.16 |
ENST00000297913.3
|
OR1Q1
|
olfactory receptor family 1 subfamily Q member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.4 | 2.6 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.3 | 1.7 | GO:0010193 | response to ozone(GO:0010193) |
| 0.3 | 2.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.3 | 2.2 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.3 | 0.8 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.3 | 0.8 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.2 | 0.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.2 | 0.8 | GO:0052047 | positive regulation of fibrinolysis(GO:0051919) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.2 | 3.0 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.2 | 0.5 | GO:0048290 | isotype switching to IgA isotypes(GO:0048290) |
| 0.2 | 0.7 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 4.7 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.2 | 1.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.9 | GO:2001107 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.1 | 2.9 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.5 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.1 | 1.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 2.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.3 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 1.4 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.3 | GO:0020012 | evasion or tolerance of host immune response(GO:0020012) evasion or tolerance of host defense response(GO:0030682) evasion or tolerance by virus of host immune response(GO:0030683) evasion or tolerance of immune response of other organism involved in symbiotic interaction(GO:0051805) evasion or tolerance of defense response of other organism involved in symbiotic interaction(GO:0051807) |
| 0.1 | 0.6 | GO:1900220 | negative regulation of alkaline phosphatase activity(GO:0010693) semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 1.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.7 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.4 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.1 | 0.6 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.3 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 0.3 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 0.2 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.1 | 0.8 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.5 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 1.2 | GO:0042940 | serine transport(GO:0032329) D-amino acid transport(GO:0042940) |
| 0.1 | 1.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.1 | 0.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 1.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.5 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.5 | GO:0031947 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.0 | 0.6 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.4 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.3 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.4 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.9 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.0 | 0.1 | GO:0002760 | positive regulation of antimicrobial humoral response(GO:0002760) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 1.1 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.6 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.6 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.5 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 1.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 2.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.4 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.8 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 1.0 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.8 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.7 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.4 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.2 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.5 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0044279 | extrinsic component of external side of plasma membrane(GO:0031232) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 1.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.9 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 4.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.0 | 1.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 3.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.7 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 2.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 2.4 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 5.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.5 | 4.7 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.3 | 2.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.3 | 0.8 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.2 | 2.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.2 | 1.4 | GO:0019862 | IgA binding(GO:0019862) |
| 0.2 | 2.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 1.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.6 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.4 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.7 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 0.6 | GO:0005119 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.3 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 1.7 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.7 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.6 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.1 | 0.3 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 3.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.4 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 1.9 | GO:0023026 | MHC protein complex binding(GO:0023023) MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.8 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 1.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 1.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.5 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.5 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 2.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 2.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 4.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 2.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 2.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 3.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |