Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
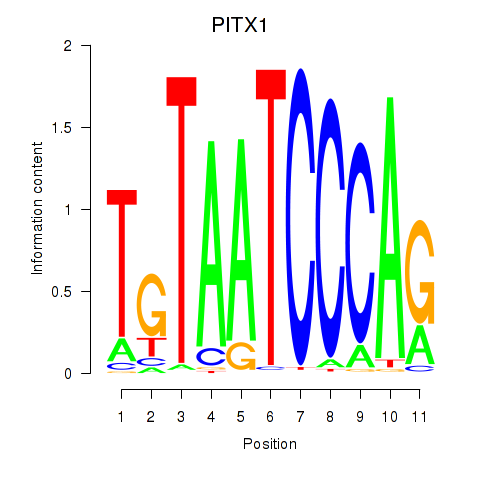
Results for PITX1
Z-value: 3.23
Transcription factors associated with PITX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX1
|
ENSG00000069011.16 | paired like homeodomain 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX1 | hg38_v1_chr5_-_135034212_135034294 | -0.04 | 8.4e-01 | Click! |
Activity profile of PITX1 motif
Sorted Z-values of PITX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 1.9 | 5.6 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 1.5 | 7.5 | GO:0032796 | uropod organization(GO:0032796) |
| 1.3 | 5.3 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 1.3 | 1.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.2 | 3.6 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 1.2 | 8.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 1.1 | 20.4 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 1.1 | 3.4 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.1 | 5.3 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.9 | 2.7 | GO:0071613 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.9 | 2.7 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.9 | 3.6 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.9 | 7.9 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.9 | 3.5 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.8 | 2.5 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) response to L-arginine(GO:1903576) |
| 0.8 | 4.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.8 | 1.5 | GO:0002339 | B cell selection(GO:0002339) |
| 0.7 | 2.2 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.7 | 2.2 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.7 | 3.7 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.7 | 5.1 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.7 | 10.8 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.7 | 0.7 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 0.7 | 5.5 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.7 | 2.7 | GO:0050992 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.7 | 1.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.7 | 4.6 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.7 | 4.6 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.6 | 6.0 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.6 | 1.7 | GO:1904826 | regulation of phenotypic switching by transcription from RNA polymerase II promoter(GO:0100057) regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.6 | 5.6 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.5 | 12.5 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.5 | 1.6 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.5 | 4.7 | GO:0051001 | negative regulation of action potential(GO:0045759) negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.5 | 3.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.5 | 1.5 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.5 | 8.2 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.5 | 2.4 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.5 | 2.8 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.5 | 1.8 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.5 | 2.3 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.5 | 2.3 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.4 | 1.8 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.4 | 1.3 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.4 | 1.8 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.4 | 2.2 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.4 | 6.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 | 3.0 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.4 | 4.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.4 | 3.4 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.4 | 1.3 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.4 | 1.7 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.4 | 7.0 | GO:0051873 | killing by host of symbiont cells(GO:0051873) |
| 0.4 | 1.2 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.4 | 1.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.4 | 2.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.4 | 1.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.4 | 2.7 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.4 | 12.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.4 | 1.5 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.4 | 2.6 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.4 | 1.5 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.4 | 2.5 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.4 | 1.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.4 | 1.1 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.4 | 1.1 | GO:0006463 | steroid hormone receptor complex assembly(GO:0006463) |
| 0.4 | 5.0 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.4 | 2.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.4 | 2.1 | GO:1990539 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.3 | 3.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.3 | 2.1 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.3 | 0.7 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.3 | 1.0 | GO:0010905 | regulation of UDP-glucose catabolic process(GO:0010904) negative regulation of UDP-glucose catabolic process(GO:0010905) regulation of glycogen synthase activity, transferring glucose-1-phosphate(GO:1904226) negative regulation of glycogen synthase activity, transferring glucose-1-phosphate(GO:1904227) |
| 0.3 | 4.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.3 | 1.0 | GO:0090410 | malonate catabolic process(GO:0090410) |
| 0.3 | 3.0 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.3 | 3.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 1.6 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.3 | 0.7 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.3 | 2.9 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.3 | 1.9 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.3 | 1.2 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.3 | 1.2 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.3 | 0.9 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.3 | 0.9 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.3 | 2.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 0.9 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.3 | 0.9 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.3 | 2.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 0.9 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.3 | 2.0 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.3 | 3.2 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.3 | 3.5 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.3 | 1.4 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.3 | 0.8 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.3 | 2.5 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.3 | 1.1 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.3 | 1.6 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.3 | 4.7 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.3 | 0.8 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of meiosis I(GO:0060903) |
| 0.3 | 1.9 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 0.5 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.3 | 0.8 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.3 | 0.8 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.3 | 2.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.3 | 1.0 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.3 | 10.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.3 | 2.8 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.3 | 3.3 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.3 | 1.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 | 8.6 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.3 | 3.8 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.3 | 11.3 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.3 | 0.8 | GO:0044349 | nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) DNA 3' dephosphorylation(GO:0098503) DNA 3' dephosphorylation involved in DNA repair(GO:0098504) polynucleotide 3' dephosphorylation(GO:0098506) |
| 0.3 | 1.5 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 9.9 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.2 | 3.2 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.2 | 2.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 1.4 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.2 | 1.9 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.2 | 7.4 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.2 | 0.7 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 1.2 | GO:1902896 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) terminal web assembly(GO:1902896) |
| 0.2 | 2.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.2 | 0.5 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.2 | 2.0 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 2.9 | GO:0006188 | IMP biosynthetic process(GO:0006188) |
| 0.2 | 6.0 | GO:0034397 | telomere localization(GO:0034397) meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.2 | 1.8 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.2 | 2.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.1 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.2 | 0.8 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.2 | 1.5 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.2 | 2.9 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 0.6 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.2 | 4.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 3.9 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.2 | 0.8 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 2.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.2 | 1.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 0.6 | GO:0090108 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.2 | 4.9 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.2 | 0.7 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 0.2 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.2 | 0.6 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.2 | 29.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.2 | 1.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 1.5 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 0.7 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.2 | 1.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 4.8 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 0.7 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.2 | 0.4 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.2 | 1.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 0.7 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.2 | 6.2 | GO:0007140 | male meiosis(GO:0007140) |
| 0.2 | 0.3 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 0.2 | 0.5 | GO:0006212 | uracil catabolic process(GO:0006212) |
| 0.2 | 2.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 1.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 1.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 1.0 | GO:0050668 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.2 | 0.2 | GO:0039519 | modulation by virus of host autophagy(GO:0039519) |
| 0.2 | 1.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 2.5 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 0.5 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.2 | 0.3 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.2 | 2.7 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 0.6 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.2 | 2.9 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.2 | 0.5 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 2.8 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.2 | 1.2 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.2 | 2.0 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.1 | 2.7 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.4 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 1.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.9 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.1 | 1.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 2.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.4 | GO:1900368 | pre-miRNA export from nucleus(GO:0035281) regulation of RNA interference(GO:1900368) |
| 0.1 | 1.0 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.6 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 3.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 1.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 1.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 3.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.7 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 1.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.8 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.9 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 2.9 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 1.5 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 5.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 4.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.0 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.8 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 1.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 1.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.4 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.6 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 5.2 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 1.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 2.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 2.9 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.1 | 1.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 0.4 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.7 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 1.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.7 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 1.4 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 1.0 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.7 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.6 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 0.7 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 1.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 1.0 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.9 | GO:0070092 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.1 | 0.3 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.6 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 1.2 | GO:2000371 | meiotic chromosome condensation(GO:0010032) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 1.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 1.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 1.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 18.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.8 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 0.3 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 1.0 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 0.4 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.3 | GO:0034473 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) U1 snRNA 3'-end processing(GO:0034473) U4 snRNA 3'-end processing(GO:0034475) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.1 | 0.5 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.9 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 2.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.4 | GO:0070781 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.1 | 0.6 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 1.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 1.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.4 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 1.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.9 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.7 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 4.5 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 1.5 | GO:0042940 | serine transport(GO:0032329) D-amino acid transport(GO:0042940) |
| 0.1 | 0.6 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.7 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.4 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.1 | 0.6 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.2 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 3.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.3 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 1.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.7 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 2.5 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 2.3 | GO:1902603 | carnitine transmembrane transport(GO:1902603) |
| 0.1 | 2.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 6.0 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 3.1 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 0.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 3.8 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.1 | 3.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 0.3 | GO:0032262 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) |
| 0.1 | 1.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.4 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.8 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 1.7 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.1 | 1.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 4.6 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.1 | 0.4 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 1.0 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.1 | 1.1 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.1 | 2.0 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 1.8 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.1 | 0.3 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.1 | 1.4 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.1 | 0.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 0.5 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.8 | GO:0071287 | cellular response to manganese ion(GO:0071287) |
| 0.1 | 0.9 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.3 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 3.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 1.2 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) |
| 0.1 | 0.3 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 1.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 2.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.6 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.1 | 4.3 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.1 | 0.2 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 0.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.7 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.5 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 1.4 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 | 0.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.4 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 3.8 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.3 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.5 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.5 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 1.8 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 3.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 1.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 3.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 1.7 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.5 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.9 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 3.5 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 0.2 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 1.3 | GO:0006309 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.9 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.8 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.3 | GO:0003068 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 2.8 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.1 | 0.4 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.1 | 0.6 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 1.9 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.1 | 1.1 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.1 | 1.2 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.1 | 0.4 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 2.7 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.1 | 1.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.8 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 0.5 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 1.1 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.9 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 2.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.2 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 2.5 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 2.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.5 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0032258 | CVT pathway(GO:0032258) |
| 0.0 | 1.4 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.6 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.4 | GO:0046498 | S-adenosylmethionine cycle(GO:0033353) S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 2.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 2.3 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.3 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.3 | GO:0002265 | astrocyte activation involved in immune response(GO:0002265) |
| 0.0 | 0.1 | GO:0071335 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 2.9 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 1.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.3 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 1.3 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 1.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 24.7 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 1.2 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 5.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.5 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 1.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.4 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.6 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.0 | 0.4 | GO:0002475 | antigen processing and presentation via MHC class Ib(GO:0002475) |
| 0.0 | 0.2 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cell differentiation(GO:0001826) inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.3 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 1.4 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 1.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.4 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 1.0 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 2.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 1.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 1.4 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.6 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 1.3 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 0.2 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 2.7 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.0 | 0.2 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 0.5 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 1.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.4 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.7 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.0 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 1.5 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.6 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 2.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.5 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 2.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.3 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 2.5 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.6 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 1.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.8 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.0 | 0.6 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.1 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.9 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.7 | GO:0048753 | melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.0 | 0.3 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.3 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.5 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 8.2 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.5 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.7 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.0 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.4 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.9 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 18.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.8 | 8.2 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.8 | 5.6 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.8 | 2.4 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.8 | 7.9 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.7 | 2.1 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.7 | 2.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.6 | 1.9 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.6 | 7.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.6 | 3.5 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.6 | 2.9 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.5 | 2.4 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.5 | 3.4 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.5 | 1.4 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.4 | 3.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.4 | 5.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.4 | 1.6 | GO:0000811 | GINS complex(GO:0000811) |
| 0.4 | 2.7 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.4 | 1.5 | GO:0097229 | sperm end piece(GO:0097229) |
| 0.3 | 1.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.3 | 1.4 | GO:1990879 | CST complex(GO:1990879) |
| 0.3 | 3.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 2.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 1.0 | GO:0090651 | apical cytoplasm(GO:0090651) |
| 0.3 | 13.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 10.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.3 | 1.9 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.3 | 1.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.3 | 9.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 1.7 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.3 | 5.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 1.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 0.8 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.3 | 2.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.3 | 1.8 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 1.6 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 2.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 1.9 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 2.5 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 1.5 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 4.0 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 24.5 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.2 | 1.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.2 | 1.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 5.0 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 1.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 2.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 1.3 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 3.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 2.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 2.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 10.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 1.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 1.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 9.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 2.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 4.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.4 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.1 | 0.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 3.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.7 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 6.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 5.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 5.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.4 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.1 | 0.4 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.1 | 1.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 1.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 1.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 9.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.9 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 1.2 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 14.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 1.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 4.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 10.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 1.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 10.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 2.2 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 0.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 3.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 5.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.8 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 3.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 1.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.1 | 0.8 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.5 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 1.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.9 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 1.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.7 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 2.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 1.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 5.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 7.5 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 2.0 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 3.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 8.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 2.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 6.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 1.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.7 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 2.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:1905103 | integral component of lysosomal membrane(GO:1905103) |
| 0.0 | 0.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 3.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 3.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 1.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 3.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 1.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 7.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 1.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 3.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 1.2 | 4.7 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 1.1 | 1.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 1.1 | 5.5 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.0 | 4.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.9 | 2.7 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.9 | 2.7 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.9 | 3.5 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.9 | 5.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.8 | 2.5 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.8 | 2.5 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.8 | 5.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.8 | 7.9 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.8 | 5.5 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.7 | 8.2 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.7 | 8.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.7 | 2.7 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.6 | 1.9 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.6 | 1.9 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.6 | 4.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.6 | 4.5 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.6 | 5.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.6 | 2.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.5 | 2.6 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.5 | 4.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.5 | 3.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.5 | 2.4 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.5 | 2.3 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.5 | 5.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.5 | 1.8 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.5 | 3.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.4 | 1.3 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.4 | 3.9 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.4 | 1.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.4 | 6.0 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.4 | 3.4 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.4 | 0.4 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.4 | 4.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.4 | 2.8 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.4 | 0.8 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.4 | 1.2 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.4 | 1.6 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.4 | 3.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.4 | 1.9 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.4 | 1.9 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.4 | 3.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.4 | 4.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.4 | 1.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.4 | 8.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 1.4 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.4 | 2.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.3 | 1.7 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.3 | 5.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 2.0 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.3 | 1.4 | GO:0002046 | opsin binding(GO:0002046) |
| 0.3 | 1.0 | GO:0090409 | malonyl-CoA synthetase activity(GO:0090409) |
| 0.3 | 2.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.3 | 2.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.3 | 2.3 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.3 | 1.9 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.3 | 5.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 1.7 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.3 | 0.8 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 0.8 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.3 | 2.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.3 | 0.8 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.3 | 0.8 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.2 | 0.7 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.2 | 1.5 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 1.0 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.2 | 1.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.2 | 1.9 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 3.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 0.6 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 7.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 1.2 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 0.6 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.2 | 3.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.9 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 3.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 2.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 0.6 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.2 | 1.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 1.8 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 1.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 1.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 3.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 4.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 0.7 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 2.8 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.2 | 2.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 11.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 0.7 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 1.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 11.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 1.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.2 | 1.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 1.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 2.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.2 | 1.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 2.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 2.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 1.7 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 0.9 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 4.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.3 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 2.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.4 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 4.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 2.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.6 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 1.4 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 2.6 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 1.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 1.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 1.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.4 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 1.9 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.7 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.1 | 1.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 2.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.9 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.5 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 1.6 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 0.8 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.1 | 2.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 5.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 3.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 5.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 1.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.3 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 4.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 2.1 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 1.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.3 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.7 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 0.5 | GO:0002054 | nucleobase binding(GO:0002054) dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.3 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 0.3 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.1 | 0.7 | GO:0005119 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.1 | 3.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 3.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.9 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 1.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.5 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.7 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 6.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 7.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 4.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.4 | GO:0018271 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.1 | 1.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 5.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 3.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.7 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 5.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 2.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 1.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 5.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 3.5 | GO:0043028 | cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 1.0 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.0 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.1 | 1.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.4 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 2.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.1 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 1.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 15.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.5 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.7 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 3.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.2 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.1 | 0.6 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.8 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 1.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 2.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.6 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 1.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.7 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 1.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.2 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 2.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 3.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 5.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 4.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 2.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.3 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.1 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.6 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.1 | 0.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 4.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.3 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 3.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0019785 | ISG15-specific protease activity(GO:0019785) |
| 0.0 | 0.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.7 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 1.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 1.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 1.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.3 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 1.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.8 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 1.3 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.6 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.0 | 1.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.6 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.8 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 5.8 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 1.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 4.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 1.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.2 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 4.7 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 13.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.3 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 1.7 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 3.3 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 4.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.6 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 3.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 14.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 2.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 4.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 2.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.4 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 2.1 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 9.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 9.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.0 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 5.6 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.9 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 1.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.4 | 26.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 3.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 10.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 13.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 8.8 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 5.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.6 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 9.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.6 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 11.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 6.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.8 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 3.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 1.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 1.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 6.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 6.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 6.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 11.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 1.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 3.2 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 3.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 2.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.6 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 3.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 6.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 4.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 3.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 7.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 19.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 5.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 2.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.5 | 7.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 7.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 7.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.3 | 6.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 6.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 7.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 4.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 10.8 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.2 | 5.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 4.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 0.8 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.2 | 3.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 2.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 5.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 1.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 4.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 4.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 2.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 3.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 4.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 4.8 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 1.9 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 3.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 14.3 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.1 | 4.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 9.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 4.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 2.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 13.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 5.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 10.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 4.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 9.4 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 2.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.9 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 2.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 2.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 9.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 3.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 3.5 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 1.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 3.4 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.1 | 3.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 3.9 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.1 | 3.1 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 2.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 3.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 16.2 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 3.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 4.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 1.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 2.4 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 2.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.5 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.1 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 2.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 2.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 2.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.8 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 1.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |