Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
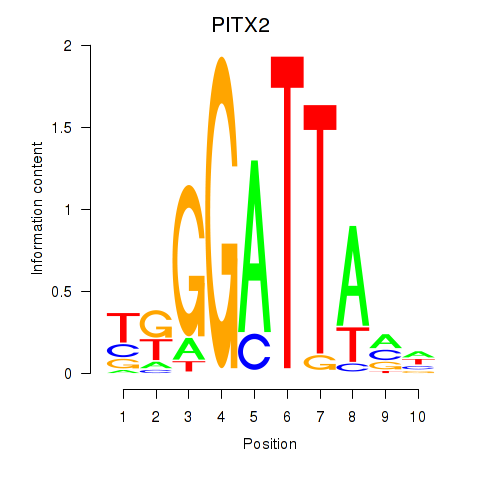
Results for PITX2
Z-value: 0.55
Transcription factors associated with PITX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX2
|
ENSG00000164093.17 | paired like homeodomain 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX2 | hg38_v1_chr4_-_110641920_110642123 | 0.05 | 7.9e-01 | Click! |
Activity profile of PITX2 motif
Sorted Z-values of PITX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_5227063 | 4.64 |
ENST00000335295.4
ENST00000485743.1 ENST00000647020.1 |
HBB
|
hemoglobin subunit beta |
| chr17_-_41518878 | 1.85 |
ENST00000254043.8
|
KRT15
|
keratin 15 |
| chr9_-_92404559 | 1.41 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chr9_+_34458752 | 1.37 |
ENST00000614641.4
ENST00000242317.9 ENST00000437363.5 |
DNAI1
|
dynein axonemal intermediate chain 1 |
| chr13_-_35855627 | 1.26 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr13_-_35855758 | 1.01 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr15_+_40929338 | 0.93 |
ENST00000249749.7
|
DLL4
|
delta like canonical Notch ligand 4 |
| chr3_-_49799821 | 0.83 |
ENST00000343366.8
ENST00000487256.1 ENST00000412678.7 |
CDHR4
|
cadherin related family member 4 |
| chr10_-_48274567 | 0.83 |
ENST00000636244.1
ENST00000374201.8 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr11_+_61755372 | 0.82 |
ENST00000265460.9
|
MYRF
|
myelin regulatory factor |
| chr11_-_119346695 | 0.80 |
ENST00000619721.6
|
MFRP
|
membrane frizzled-related protein |
| chr13_+_73058993 | 0.78 |
ENST00000377687.6
|
KLF5
|
Kruppel like factor 5 |
| chr3_+_159069252 | 0.64 |
ENST00000640015.1
ENST00000476809.7 ENST00000485419.7 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr9_+_127678455 | 0.60 |
ENST00000494254.3
|
STXBP1
|
syntaxin binding protein 1 |
| chr1_+_43300971 | 0.59 |
ENST00000372476.8
ENST00000538015.1 |
TIE1
|
tyrosine kinase with immunoglobulin like and EGF like domains 1 |
| chr3_+_154121366 | 0.56 |
ENST00000465093.6
ENST00000496710.5 ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor 26 |
| chrX_-_6228835 | 0.55 |
ENST00000381095.8
|
NLGN4X
|
neuroligin 4 X-linked |
| chr16_-_67980483 | 0.54 |
ENST00000268793.6
ENST00000672962.1 |
DPEP3
|
dipeptidase 3 |
| chr7_+_74453790 | 0.51 |
ENST00000265755.7
ENST00000424337.7 ENST00000455841.6 |
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr20_+_45722329 | 0.49 |
ENST00000279058.4
|
SPINT4
|
serine peptidase inhibitor, Kunitz type 4 |
| chr1_-_165445220 | 0.44 |
ENST00000619224.1
|
RXRG
|
retinoid X receptor gamma |
| chr10_+_102226293 | 0.44 |
ENST00000370005.4
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr4_-_101347551 | 0.43 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr16_+_28711417 | 0.42 |
ENST00000395587.5
ENST00000569690.5 ENST00000331666.11 ENST00000564243.5 ENST00000566866.5 |
EIF3C
|
eukaryotic translation initiation factor 3 subunit C |
| chr4_-_101347327 | 0.42 |
ENST00000394853.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr4_-_101346842 | 0.41 |
ENST00000507176.5
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr2_-_65432591 | 0.40 |
ENST00000356388.9
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr2_-_65432628 | 0.39 |
ENST00000440972.1
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr1_-_165445088 | 0.39 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma |
| chr4_-_101347492 | 0.39 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr1_+_91952162 | 0.39 |
ENST00000402388.1
ENST00000680541.1 |
BRDT
|
bromodomain testis associated |
| chr12_+_56521798 | 0.38 |
ENST00000262031.10
|
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr9_-_14300231 | 0.37 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B |
| chr10_+_17951906 | 0.36 |
ENST00000377371.3
ENST00000377369.7 |
SLC39A12
|
solute carrier family 39 member 12 |
| chr11_-_133532493 | 0.35 |
ENST00000524381.6
|
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr7_-_122699108 | 0.35 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133 |
| chr4_-_101347471 | 0.34 |
ENST00000323055.10
ENST00000512215.5 |
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr6_+_46693835 | 0.33 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr1_+_202010575 | 0.32 |
ENST00000367283.7
ENST00000367284.10 |
ELF3
|
E74 like ETS transcription factor 3 |
| chr10_+_17951825 | 0.32 |
ENST00000539911.5
|
SLC39A12
|
solute carrier family 39 member 12 |
| chr16_-_11976611 | 0.32 |
ENST00000538896.5
ENST00000673243.1 |
NPIPB2
|
nuclear pore complex interacting protein family member B2 |
| chr12_-_89656093 | 0.31 |
ENST00000359142.7
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr10_+_17951885 | 0.30 |
ENST00000377374.8
|
SLC39A12
|
solute carrier family 39 member 12 |
| chr12_-_89656051 | 0.27 |
ENST00000261173.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr9_-_107489754 | 0.24 |
ENST00000610832.1
ENST00000374672.5 |
KLF4
|
Kruppel like factor 4 |
| chr5_-_150758497 | 0.23 |
ENST00000521533.1
ENST00000424236.5 |
DCTN4
|
dynactin subunit 4 |
| chr17_-_4739866 | 0.23 |
ENST00000574412.6
ENST00000293778.12 |
CXCL16
|
C-X-C motif chemokine ligand 16 |
| chr8_+_48008409 | 0.19 |
ENST00000523432.5
ENST00000521346.5 ENST00000523111.7 ENST00000517630.5 |
UBE2V2
|
ubiquitin conjugating enzyme E2 V2 |
| chr5_+_14441043 | 0.19 |
ENST00000639876.2
|
TRIO
|
trio Rho guanine nucleotide exchange factor |
| chr6_+_31137646 | 0.17 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr22_-_30471986 | 0.17 |
ENST00000401751.5
ENST00000402286.5 ENST00000403066.5 ENST00000215812.9 |
SEC14L3
|
SEC14 like lipid binding 3 |
| chr17_+_4740042 | 0.12 |
ENST00000592813.5
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr1_+_202010615 | 0.11 |
ENST00000446188.1
|
ELF3
|
E74 like ETS transcription factor 3 |
| chr11_-_96343170 | 0.10 |
ENST00000524717.6
|
MAML2
|
mastermind like transcriptional coactivator 2 |
| chr17_+_4740005 | 0.09 |
ENST00000269289.10
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr12_+_40393392 | 0.09 |
ENST00000676020.1
|
MUC19
|
mucin 19, oligomeric |
| chr5_+_38258373 | 0.09 |
ENST00000354891.7
|
EGFLAM
|
EGF like, fibronectin type III and laminin G domains |
| chr1_+_2476315 | 0.06 |
ENST00000419816.6
|
PLCH2
|
phospholipase C eta 2 |
| chr12_+_69739370 | 0.05 |
ENST00000550536.5
ENST00000362025.9 |
RAB3IP
|
RAB3A interacting protein |
| chr6_+_130018565 | 0.05 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr19_+_2476118 | 0.03 |
ENST00000215631.9
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA damage inducible beta |
| chr16_-_15642488 | 0.02 |
ENST00000549219.1
|
MARF1
|
meiosis regulator and mRNA stability factor 1 |
| chr1_+_2476284 | 0.02 |
ENST00000378486.8
|
PLCH2
|
phospholipase C eta 2 |
| chr13_-_94479671 | 0.01 |
ENST00000377028.10
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr6_+_35342614 | 0.01 |
ENST00000337400.6
ENST00000311565.4 |
PPARD
|
peroxisome proliferator activated receptor delta |
| chr6_-_159788439 | 0.01 |
ENST00000539948.5
|
TCP1
|
t-complex 1 |
| chr6_-_111483700 | 0.00 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr21_-_18403754 | 0.00 |
ENST00000284885.8
|
TMPRSS15
|
transmembrane serine protease 15 |
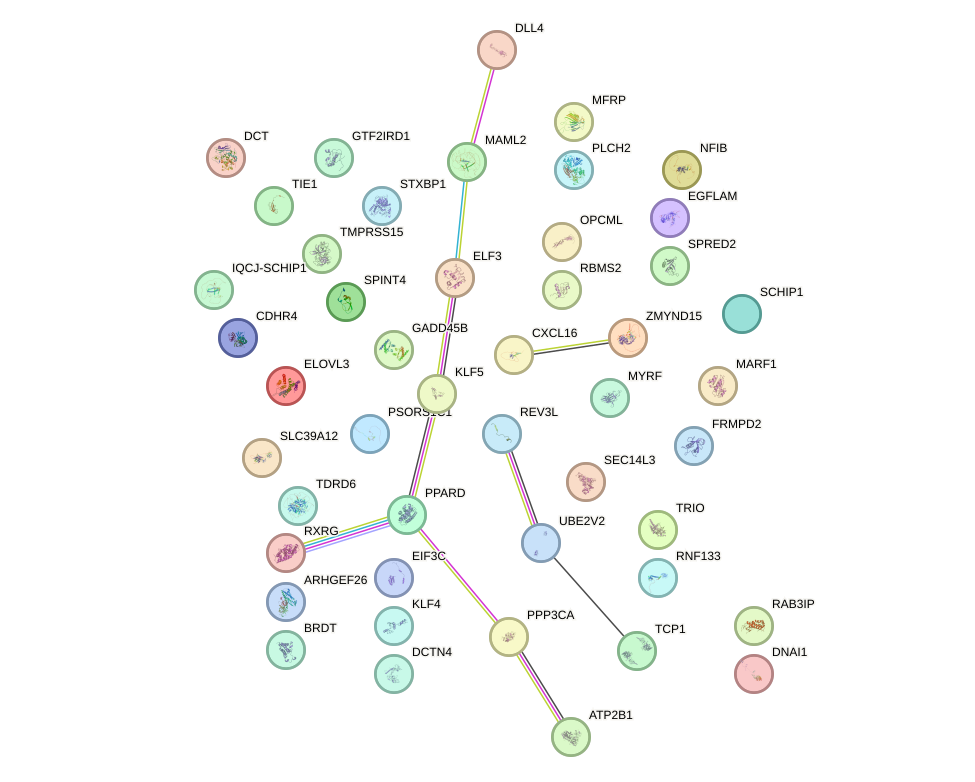
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.6 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.4 | 2.0 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 0.9 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.4 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.5 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.6 | GO:1903294 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.1 | 1.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.6 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 0.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 1.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.2 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 1.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.6 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 2.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.4 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.6 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.4 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.8 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 2.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 0.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.8 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 2.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 1.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0043274 | phospholipase binding(GO:0043274) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.9 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 2.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 4.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |