Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for POU5F1_POU2F3
Z-value: 4.92
Transcription factors associated with POU5F1_POU2F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
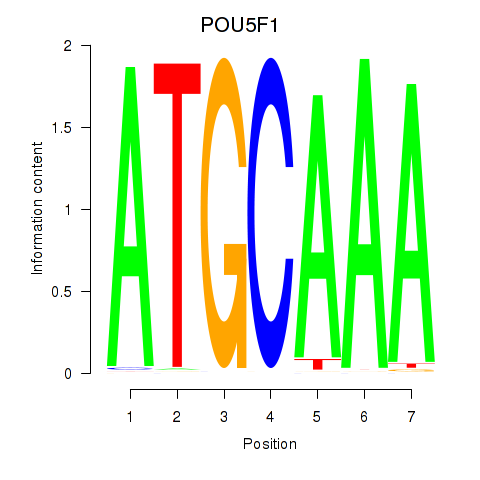
POU5F1
|
ENSG00000204531.20 | POU class 5 homeobox 1 |
|
POU2F3
|
ENSG00000137709.10 | POU class 2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU5F1 | hg38_v1_chr6_-_31170620_31170698 | 0.17 | 3.6e-01 | Click! |
| POU2F3 | hg38_v1_chr11_+_120236635_120236642 | 0.05 | 7.7e-01 | Click! |
Activity profile of POU5F1_POU2F3 motif
Sorted Z-values of POU5F1_POU2F3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_22720615 | 38.34 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr14_-_106593319 | 30.13 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr22_+_22686724 | 28.88 |
ENST00000390305.2
|
IGLV3-25
|
immunoglobulin lambda variable 3-25 |
| chr14_-_106117159 | 27.44 |
ENST00000390601.3
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 |
| chr22_+_22409755 | 27.31 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr2_+_89959979 | 27.22 |
ENST00000453166.2
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr2_-_88947820 | 25.86 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr2_-_89100352 | 25.41 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr14_-_106277039 | 25.36 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr22_+_22357739 | 25.11 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr22_+_22758698 | 24.94 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr2_-_89268506 | 24.82 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr2_+_89913982 | 24.50 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr14_-_105987068 | 24.29 |
ENST00000390594.3
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr2_-_89245596 | 23.87 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr14_-_106675544 | 23.43 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr2_+_89936859 | 23.22 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr14_-_106154113 | 23.07 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr14_-_106803221 | 22.77 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr14_-_106715166 | 22.73 |
ENST00000390633.2
|
IGHV1-69
|
immunoglobulin heavy variable 1-69 |
| chr2_+_90114838 | 22.72 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 |
| chr22_+_22195753 | 22.35 |
ENST00000390285.4
|
IGLV6-57
|
immunoglobulin lambda variable 6-57 |
| chr14_-_106579223 | 22.35 |
ENST00000390626.2
|
IGHV5-51
|
immunoglobulin heavy variable 5-51 |
| chr2_+_90100235 | 22.20 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chrX_-_48919015 | 21.70 |
ENST00000376509.4
|
PIM2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr2_-_89143133 | 21.45 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20 |
| chr2_-_89040745 | 21.27 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr2_-_89222461 | 21.25 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr14_-_106762576 | 21.11 |
ENST00000624687.1
|
IGHV1-69D
|
immunoglobulin heavy variable 1-69D |
| chr2_+_90159840 | 20.35 |
ENST00000377032.5
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr14_-_106360320 | 20.29 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr22_+_22098683 | 20.25 |
ENST00000390283.2
|
IGLV8-61
|
immunoglobulin lambda variable 8-61 |
| chr2_+_90038848 | 20.02 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr2_-_88992903 | 19.65 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr22_+_22030934 | 19.51 |
ENST00000390282.2
|
IGLV4-69
|
immunoglobulin lambda variable 4-69 |
| chr14_-_106130061 | 19.50 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr2_-_89177160 | 19.20 |
ENST00000484817.1
|
IGKV2-24
|
immunoglobulin kappa variable 2-24 |
| chr2_+_89947508 | 19.15 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr14_-_106627685 | 19.04 |
ENST00000390629.3
|
IGHV4-59
|
immunoglobulin heavy variable 4-59 |
| chr4_-_70666492 | 18.85 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr22_+_22792485 | 18.78 |
ENST00000390314.2
|
IGLV2-11
|
immunoglobulin lambda variable 2-11 |
| chr22_+_22880706 | 18.50 |
ENST00000390319.2
|
IGLV3-1
|
immunoglobulin lambda variable 3-1 |
| chr2_+_88885397 | 18.36 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr14_-_106538331 | 18.08 |
ENST00000390624.3
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr2_+_90154073 | 17.77 |
ENST00000611391.1
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr14_-_106374129 | 17.74 |
ENST00000390616.2
|
IGHV4-34
|
immunoglobulin heavy variable 4-34 |
| chr14_-_106012390 | 17.18 |
ENST00000455737.2
|
IGHV4-4
|
immunoglobulin heavy variable 4-4 |
| chr22_+_22431949 | 17.06 |
ENST00000390301.3
|
IGLV1-36
|
immunoglobulin lambda variable 1-36 |
| chr22_+_22734577 | 16.61 |
ENST00000390310.3
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr22_+_22747383 | 16.50 |
ENST00000390311.3
|
IGLV3-16
|
immunoglobulin lambda variable 3-16 |
| chr16_+_32066065 | 16.18 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr22_+_22818994 | 16.02 |
ENST00000390316.2
|
IGLV3-9
|
immunoglobulin lambda variable 3-9 |
| chr2_-_89320146 | 15.88 |
ENST00000498574.1
|
IGKV1-39
|
immunoglobulin kappa variable 1-39 |
| chr14_-_106470788 | 15.50 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr22_+_22668286 | 15.39 |
ENST00000390304.2
|
IGLV3-27
|
immunoglobulin lambda variable 3-27 |
| chr2_+_90004792 | 15.14 |
ENST00000462693.1
|
IGKV2D-24
|
immunoglobulin kappa variable 2D-24 (non-functional) |
| chr14_-_106108453 | 14.96 |
ENST00000632950.2
|
IGHV5-10-1
|
immunoglobulin heavy variable 5-10-1 |
| chr14_-_106088573 | 14.68 |
ENST00000632099.1
|
IGHV3-64D
|
immunoglobulin heavy variable 3-64D |
| chr2_-_89160329 | 14.31 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr4_-_70666961 | 14.19 |
ENST00000510437.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr15_-_21742799 | 14.04 |
ENST00000622410.2
|
ENSG00000278263.2
|
novel protein, identical to IGHV4-4 |
| chr2_+_90082635 | 14.03 |
ENST00000483379.1
|
IGKV1D-17
|
immunoglobulin kappa variable 1D-17 |
| chr22_+_22395005 | 13.75 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr2_+_89851723 | 13.65 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr22_+_22822658 | 13.59 |
ENST00000620395.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr22_+_22380766 | 13.37 |
ENST00000390297.3
|
IGLV1-44
|
immunoglobulin lambda variable 1-44 |
| chr2_+_89862438 | 13.25 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr2_+_88897230 | 13.18 |
ENST00000390244.2
|
IGKV5-2
|
immunoglobulin kappa variable 5-2 |
| chr4_-_70666884 | 13.09 |
ENST00000510614.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr14_-_106622837 | 12.92 |
ENST00000390628.3
|
IGHV1-58
|
immunoglobulin heavy variable 1-58 |
| chr6_-_32816910 | 12.91 |
ENST00000447394.1
ENST00000438763.7 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr14_-_106791226 | 12.65 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr2_-_181657096 | 12.62 |
ENST00000410087.8
ENST00000409440.7 |
CERKL
|
ceramide kinase like |
| chr2_+_90172802 | 12.50 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr2_-_89117844 | 12.34 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr2_+_90021567 | 12.20 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr14_-_106335613 | 12.12 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr2_-_89085787 | 11.94 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr2_-_89213917 | 11.92 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr22_+_22811737 | 11.86 |
ENST00000390315.3
|
IGLV3-10
|
immunoglobulin lambda variable 3-10 |
| chr14_-_106062670 | 11.78 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr22_+_22697789 | 11.67 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr14_-_106038355 | 11.37 |
ENST00000390597.3
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr14_-_106422175 | 11.37 |
ENST00000390619.2
|
IGHV4-39
|
immunoglobulin heavy variable 4-39 |
| chr2_+_87338511 | 11.05 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr14_-_106658251 | 10.59 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr4_+_40192949 | 10.59 |
ENST00000507851.5
ENST00000615577.4 ENST00000613272.4 |
RHOH
|
ras homolog family member H |
| chr2_+_113406368 | 10.34 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr2_-_88966767 | 10.33 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr16_+_23835946 | 10.32 |
ENST00000321728.12
ENST00000643927.1 |
PRKCB
|
protein kinase C beta |
| chr2_+_90234809 | 10.29 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr7_-_3043838 | 9.79 |
ENST00000356408.3
ENST00000396946.9 |
CARD11
|
caspase recruitment domain family member 11 |
| chr14_-_106811131 | 9.76 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr14_-_106557465 | 9.68 |
ENST00000390625.3
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr2_-_88857582 | 9.41 |
ENST00000390237.2
|
IGKC
|
immunoglobulin kappa constant |
| chr14_-_106771020 | 9.40 |
ENST00000617374.2
|
IGHV2-70
|
immunoglobulin heavy variable 2-70 |
| chr9_-_34691204 | 9.27 |
ENST00000378800.3
ENST00000311925.7 |
CCL19
|
C-C motif chemokine ligand 19 |
| chr16_+_32995048 | 9.24 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr14_-_106724093 | 9.15 |
ENST00000390634.3
|
IGHV2-70D
|
immunoglobulin heavy variable 2-70D |
| chr6_-_31592992 | 9.13 |
ENST00000340027.10
|
NCR3
|
natural cytotoxicity triggering receptor 3 |
| chr2_+_89884740 | 9.07 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr19_-_38617912 | 8.99 |
ENST00000591517.5
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr22_-_23754376 | 8.95 |
ENST00000398465.3
ENST00000248948.4 |
VPREB3
|
V-set pre-B cell surrogate light chain 3 |
| chr19_+_14583076 | 8.88 |
ENST00000547437.5
ENST00000417570.6 |
CLEC17A
|
C-type lectin domain containing 17A |
| chr13_-_99307387 | 8.88 |
ENST00000376414.5
|
GPR183
|
G protein-coupled receptor 183 |
| chr16_+_33827140 | 8.80 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr6_-_107824294 | 8.77 |
ENST00000369020.8
ENST00000369022.6 |
SCML4
|
Scm polycomb group protein like 4 |
| chr22_+_22369601 | 8.64 |
ENST00000390295.3
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 |
| chr22_+_22375984 | 8.60 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr22_+_22343185 | 8.56 |
ENST00000427632.2
|
IGLV9-49
|
immunoglobulin lambda variable 9-49 |
| chr12_-_122716790 | 8.44 |
ENST00000528880.3
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr7_-_142813394 | 8.42 |
ENST00000417977.2
|
TRBV30
|
T cell receptor beta variable 30 |
| chr22_+_22322452 | 8.30 |
ENST00000390290.3
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr3_+_46354072 | 8.28 |
ENST00000445132.3
ENST00000421659.1 |
CCR2
|
C-C motif chemokine receptor 2 |
| chr14_-_106639589 | 8.25 |
ENST00000390630.3
|
IGHV4-61
|
immunoglobulin heavy variable 4-61 |
| chr16_+_33009175 | 8.23 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr14_-_106389858 | 8.19 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr14_+_21712313 | 8.15 |
ENST00000390424.2
|
TRAV2
|
T cell receptor alpha variable 2 |
| chr19_-_38617928 | 8.15 |
ENST00000396857.7
ENST00000586296.5 |
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr2_-_88979016 | 8.14 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr5_+_157180816 | 8.03 |
ENST00000422843.8
|
ITK
|
IL2 inducible T cell kinase |
| chr4_+_70226116 | 8.00 |
ENST00000317987.6
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr2_+_90209873 | 7.93 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr14_-_106211453 | 7.92 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr14_-_106301848 | 7.88 |
ENST00000390611.2
|
IGHV2-26
|
immunoglobulin heavy variable 2-26 |
| chr22_+_22162155 | 7.84 |
ENST00000390284.2
|
IGLV4-60
|
immunoglobulin lambda variable 4-60 |
| chr2_-_89297785 | 7.79 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr2_-_89010515 | 7.63 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr14_-_106025628 | 7.62 |
ENST00000631943.1
|
IGHV7-4-1
|
immunoglobulin heavy variable 7-4-1 |
| chr17_-_63932261 | 7.59 |
ENST00000349817.2
ENST00000006750.8 ENST00000392795.7 |
CD79B
|
CD79b molecule |
| chr1_+_27342014 | 7.28 |
ENST00000618673.4
ENST00000318074.9 ENST00000616558.5 |
SYTL1
|
synaptotagmin like 1 |
| chr2_-_112836702 | 6.99 |
ENST00000416750.1
ENST00000263341.7 ENST00000418817.5 |
IL1B
|
interleukin 1 beta |
| chr8_-_133102477 | 6.97 |
ENST00000522119.5
ENST00000523610.5 ENST00000338087.10 ENST00000521302.5 ENST00000519558.5 ENST00000519747.5 ENST00000517648.5 |
SLA
|
Src like adaptor |
| chr8_+_55879818 | 6.93 |
ENST00000520220.6
ENST00000519728.6 |
LYN
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr1_+_5992639 | 6.86 |
ENST00000666163.1
ENST00000671676.1 ENST00000658883.1 ENST00000445501.6 ENST00000668706.1 ENST00000653262.1 ENST00000389632.9 ENST00000341524.6 ENST00000462676.3 |
KCNAB2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr21_-_10649835 | 6.77 |
ENST00000622028.1
|
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr1_-_157777124 | 6.68 |
ENST00000361516.8
ENST00000368181.4 |
FCRL2
|
Fc receptor like 2 |
| chr14_-_106235582 | 6.60 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr14_-_106349792 | 6.59 |
ENST00000438142.3
|
IGHV4-31
|
immunoglobulin heavy variable 4-31 |
| chr19_+_35329161 | 6.56 |
ENST00000341773.10
ENST00000600131.5 ENST00000595780.5 ENST00000597916.5 ENST00000593867.5 ENST00000600424.5 ENST00000599811.5 ENST00000536635.6 ENST00000085219.10 ENST00000544992.6 ENST00000419549.6 |
CD22
|
CD22 molecule |
| chr22_+_22427517 | 6.48 |
ENST00000390300.2
|
IGLV5-37
|
immunoglobulin lambda variable 5-37 |
| chr5_-_111756245 | 6.38 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr6_-_127900958 | 6.34 |
ENST00000434358.3
ENST00000630369.2 ENST00000368248.4 |
THEMIS
|
thymocyte selection associated |
| chr6_-_31592952 | 6.23 |
ENST00000376073.8
ENST00000376072.7 |
NCR3
|
natural cytotoxicity triggering receptor 3 |
| chr2_+_90220727 | 6.18 |
ENST00000471857.2
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr17_-_75844334 | 6.11 |
ENST00000592386.5
ENST00000412096.6 ENST00000586147.1 ENST00000207549.9 |
UNC13D
|
unc-13 homolog D |
| chrX_-_13817027 | 6.11 |
ENST00000493677.5
ENST00000355135.6 ENST00000316715.9 |
GPM6B
|
glycoprotein M6B |
| chr19_-_18397596 | 5.92 |
ENST00000595840.1
ENST00000339007.4 |
LRRC25
|
leucine rich repeat containing 25 |
| chr14_-_106165730 | 5.92 |
ENST00000390604.2
|
IGHV3-16
|
immunoglobulin heavy variable 3-16 (non-functional) |
| chr17_+_14301069 | 5.83 |
ENST00000360954.3
|
HS3ST3B1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
| chr9_-_35618367 | 5.64 |
ENST00000378431.5
ENST00000378430.3 ENST00000259633.9 |
CD72
|
CD72 molecule |
| chr4_-_163613505 | 5.59 |
ENST00000339875.9
|
MARCHF1
|
membrane associated ring-CH-type finger 1 |
| chr2_+_89985922 | 5.53 |
ENST00000390268.2
|
IGKV2D-26
|
immunoglobulin kappa variable 2D-26 |
| chr1_-_157820060 | 5.52 |
ENST00000491942.1
ENST00000358292.7 |
FCRL1
|
Fc receptor like 1 |
| chr6_+_35297809 | 5.52 |
ENST00000316637.7
|
DEF6
|
DEF6 guanine nucleotide exchange factor |
| chr6_+_26124161 | 5.48 |
ENST00000377791.4
ENST00000602637.1 |
H2AC6
|
H2A clustered histone 6 |
| chr5_-_67196791 | 5.36 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr7_+_50308672 | 5.24 |
ENST00000439701.2
ENST00000438033.5 ENST00000492782.6 |
IKZF1
|
IKAROS family zinc finger 1 |
| chr6_-_154246850 | 5.21 |
ENST00000517438.5
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr8_-_133102623 | 5.04 |
ENST00000524345.5
|
SLA
|
Src like adaptor |
| chr14_-_106411021 | 5.02 |
ENST00000390618.2
|
IGHV3-38
|
immunoglobulin heavy variable 3-38 (non-functional) |
| chr6_-_27146841 | 4.98 |
ENST00000356950.2
|
H2BC12
|
H2B clustered histone 12 |
| chr15_-_19988117 | 4.94 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr5_+_55102635 | 4.93 |
ENST00000274306.7
|
GZMA
|
granzyme A |
| chr3_+_98531965 | 4.93 |
ENST00000284311.5
|
GPR15
|
G protein-coupled receptor 15 |
| chr11_-_64744102 | 4.88 |
ENST00000431822.5
ENST00000394432.8 ENST00000377486.7 |
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr1_+_192809031 | 4.85 |
ENST00000235382.7
|
RGS2
|
regulator of G protein signaling 2 |
| chr1_-_157820113 | 4.83 |
ENST00000368176.8
|
FCRL1
|
Fc receptor like 1 |
| chr1_-_157700738 | 4.82 |
ENST00000368186.9
ENST00000496769.1 ENST00000368184.8 |
FCRL3
|
Fc receptor like 3 |
| chr11_-_64744317 | 4.79 |
ENST00000419843.1
ENST00000394430.5 |
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr6_-_11778781 | 4.78 |
ENST00000414691.8
ENST00000229583.9 |
ADTRP
|
androgen dependent TFPI regulating protein |
| chr16_+_31355215 | 4.76 |
ENST00000562522.2
|
ITGAX
|
integrin subunit alpha X |
| chr6_+_12716801 | 4.76 |
ENST00000674595.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr16_+_31355165 | 4.68 |
ENST00000562918.5
ENST00000268296.9 |
ITGAX
|
integrin subunit alpha X |
| chr12_-_122703346 | 4.67 |
ENST00000328880.6
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr4_-_89307732 | 4.64 |
ENST00000609438.2
|
GPRIN3
|
GPRIN family member 3 |
| chr3_+_32951636 | 4.64 |
ENST00000330953.6
|
CCR4
|
C-C motif chemokine receptor 4 |
| chr14_-_106324743 | 4.62 |
ENST00000390612.3
|
IGHV4-28
|
immunoglobulin heavy variable 4-28 |
| chr7_-_84194781 | 4.59 |
ENST00000265362.9
|
SEMA3A
|
semaphorin 3A |
| chr1_-_153390976 | 4.58 |
ENST00000368732.5
ENST00000368733.4 |
S100A8
|
S100 calcium binding protein A8 |
| chr11_-_102874974 | 4.57 |
ENST00000571244.3
|
MMP12
|
matrix metallopeptidase 12 |
| chr22_+_22771791 | 4.51 |
ENST00000390313.3
|
IGLV3-12
|
immunoglobulin lambda variable 3-12 |
| chr1_+_206557157 | 4.50 |
ENST00000577571.5
|
RASSF5
|
Ras association domain family member 5 |
| chr8_-_133102874 | 4.50 |
ENST00000395352.7
|
SLA
|
Src like adaptor |
| chr16_-_21652598 | 4.50 |
ENST00000569602.1
ENST00000268389.6 |
IGSF6
|
immunoglobulin superfamily member 6 |
| chr13_-_99258366 | 4.46 |
ENST00000397470.5
ENST00000397473.7 |
GPR18
|
G protein-coupled receptor 18 |
| chr11_-_47378479 | 4.42 |
ENST00000533968.1
|
SPI1
|
Spi-1 proto-oncogene |
| chr6_-_26216673 | 4.41 |
ENST00000541790.3
|
H2BC8
|
H2B clustered histone 8 |
| chr2_-_207165923 | 4.41 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr11_-_47378527 | 4.40 |
ENST00000378538.8
|
SPI1
|
Spi-1 proto-oncogene |
| chr1_-_228457855 | 4.38 |
ENST00000366695.3
|
H2AW
|
H2A.W histone |
| chr5_-_141651376 | 4.37 |
ENST00000522783.5
ENST00000519800.1 ENST00000435817.7 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr16_-_88941801 | 4.35 |
ENST00000569464.5
ENST00000569443.5 |
CBFA2T3
|
CBFA2/RUNX1 partner transcriptional co-repressor 3 |
| chr15_-_19965101 | 4.33 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr4_-_38804783 | 4.28 |
ENST00000308979.7
ENST00000505940.1 ENST00000515861.5 |
TLR1
|
toll like receptor 1 |
| chr11_-_47378494 | 4.26 |
ENST00000533030.1
|
SPI1
|
Spi-1 proto-oncogene |
| chr12_+_92702983 | 4.25 |
ENST00000344636.6
ENST00000544406.2 |
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr4_+_40192989 | 4.18 |
ENST00000508513.5
|
RHOH
|
ras homolog family member H |
| chr19_+_21020634 | 4.17 |
ENST00000599548.5
ENST00000594110.5 ENST00000261560.10 |
ZNF430
|
zinc finger protein 430 |
| chr15_-_82571741 | 4.17 |
ENST00000562833.2
ENST00000611163.4 |
ENSG00000260836.2
CPEB1
|
novel protein cytoplasmic polyadenylation element binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU5F1_POU2F3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 1285.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 5.1 | 46.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 4.3 | 12.9 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 3.1 | 9.3 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 3.0 | 9.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 2.8 | 8.3 | GO:0002436 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
| 2.6 | 15.4 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 2.5 | 17.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 2.3 | 7.0 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 2.3 | 7.0 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 2.3 | 6.9 | GO:0070662 | mast cell proliferation(GO:0070662) |
| 2.0 | 8.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 1.9 | 444.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 1.9 | 5.6 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 1.7 | 5.1 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 1.5 | 10.3 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 1.4 | 4.3 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 1.2 | 6.2 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 1.1 | 3.3 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 1.1 | 4.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 1.0 | 8.9 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 1.0 | 6.8 | GO:0033029 | neutrophil apoptotic process(GO:0001781) regulation of neutrophil apoptotic process(GO:0033029) |
| 1.0 | 12.5 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.9 | 7.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.9 | 5.4 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.8 | 2.5 | GO:0002238 | response to molecule of fungal origin(GO:0002238) cellular response to molecule of fungal origin(GO:0071226) |
| 0.8 | 2.5 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.8 | 4.2 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.8 | 5.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.8 | 0.8 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.8 | 3.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.8 | 4.9 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.8 | 2.3 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.8 | 3.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.8 | 2.3 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.7 | 2.9 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.7 | 2.1 | GO:1990751 | positive regulation of Schwann cell migration(GO:1900149) regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.7 | 2.1 | GO:1901257 | regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of interleukin-3 production(GO:0032712) negative regulation of granulocyte colony-stimulating factor production(GO:0071656) negative regulation of macrophage colony-stimulating factor production(GO:1901257) |
| 0.7 | 6.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.7 | 7.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.6 | 0.6 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.6 | 2.6 | GO:0052361 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.6 | 9.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.6 | 4.9 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.6 | 2.4 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.6 | 1.2 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) regulation of eosinophil activation(GO:1902566) |
| 0.6 | 2.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.6 | 3.6 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.6 | 2.9 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.6 | 5.3 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.6 | 2.3 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.6 | 2.3 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.5 | 3.2 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 0.5 | 1.6 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.5 | 3.6 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.5 | 1.5 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.5 | 2.0 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.5 | 7.4 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.4 | 1.8 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.4 | 2.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.4 | 0.9 | GO:0071335 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.4 | 1.7 | GO:0018277 | protein deamination(GO:0018277) |
| 0.4 | 2.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.4 | 2.5 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.4 | 7.6 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.4 | 8.8 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.4 | 1.3 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.4 | 2.4 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.4 | 4.9 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.4 | 1.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.4 | 2.8 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.4 | 3.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.4 | 5.5 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.4 | 2.0 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.4 | 0.8 | GO:0033084 | regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.4 | 2.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 7.5 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.4 | 3.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.4 | 1.1 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.4 | 5.8 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.4 | 1.8 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.4 | 1.8 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.4 | 2.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.3 | 1.7 | GO:0061216 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.3 | 2.0 | GO:0002606 | regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.3 | 5.7 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.3 | 18.1 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.3 | 0.9 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.3 | 1.9 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 1.2 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.3 | 7.7 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.3 | 1.8 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 2.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 1.2 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.3 | 7.8 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.3 | 2.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.3 | 1.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.3 | 1.4 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.3 | 10.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.3 | 5.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 4.4 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.3 | 0.5 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.3 | 1.6 | GO:0021825 | substrate-dependent cerebral cortex tangential migration(GO:0021825) |
| 0.3 | 2.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.3 | 2.4 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.3 | 1.3 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.3 | 1.3 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) |
| 0.3 | 1.6 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.3 | 1.8 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.3 | 18.1 | GO:0007140 | male meiosis(GO:0007140) |
| 0.2 | 1.5 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.2 | 0.5 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.2 | 6.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 2.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 4.7 | GO:0003263 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) |
| 0.2 | 1.2 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.2 | 0.7 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.2 | 0.9 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 2.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 2.7 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.2 | 2.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 1.3 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.2 | 1.6 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 2.6 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.2 | 0.7 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.2 | 2.2 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.2 | 2.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.2 | 1.4 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.2 | 2.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 3.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 2.8 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 1.0 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.2 | 0.8 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 2.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 0.8 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.2 | 1.6 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.2 | 2.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 0.4 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.2 | 1.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 19.0 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.2 | 4.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 4.6 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.2 | 4.0 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.2 | 3.2 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.2 | 0.9 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.2 | 3.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.2 | 3.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 1.4 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.2 | 0.8 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.2 | 4.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 0.7 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 0.7 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.2 | 18.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 1.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 1.3 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.2 | 0.8 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.2 | 3.2 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 2.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 0.5 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.2 | 2.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 1.0 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.2 | 0.8 | GO:2000691 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.2 | 1.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.6 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.2 | 2.0 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.4 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.6 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 3.9 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 4.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.6 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.7 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.1 | 0.7 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 1.0 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.1 | 6.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 1.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.3 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.1 | 4.9 | GO:0072678 | T cell migration(GO:0072678) |
| 0.1 | 2.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 2.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.9 | GO:0046543 | development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 1.9 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 4.4 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 0.9 | GO:0002705 | positive regulation of leukocyte mediated immunity(GO:0002705) |
| 0.1 | 40.9 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 0.7 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.1 | 0.4 | GO:0098583 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.1 | 3.9 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 1.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 1.4 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.1 | 1.5 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 1.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 3.1 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.1 | 3.8 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 1.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.6 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.1 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 0.9 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 1.4 | GO:0009304 | tRNA transcription(GO:0009304) |
| 0.1 | 1.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.9 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 2.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.2 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.1 | 2.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.9 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 2.6 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.1 | 11.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 2.4 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 0.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.4 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.4 | GO:0010260 | organ senescence(GO:0010260) |
| 0.1 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 2.7 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.1 | 1.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.0 | GO:0003360 | brainstem development(GO:0003360) |
| 0.1 | 4.2 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 6.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 1.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.5 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 1.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.6 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 1.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 7.0 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 1.2 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 1.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.5 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.7 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 4.5 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 3.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 3.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.7 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 4.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 2.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.6 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.1 | 0.9 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 0.1 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.1 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 2.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.9 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 9.1 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 7.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.1 | GO:0006388 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.7 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.4 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 3.9 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 3.0 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.4 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.9 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 2.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.9 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.9 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 7.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.5 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 1.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.4 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.9 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 1.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.2 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 1.2 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.6 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.5 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 8.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.3 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.8 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.8 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.4 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.6 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.8 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 3.8 | GO:0030098 | lymphocyte differentiation(GO:0030098) |
| 0.0 | 1.0 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 1.8 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.3 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.6 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 1.1 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 2.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.3 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.5 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.3 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 7.7 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.5 | 67.6 | GO:0071756 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 6.7 | 360.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 2.0 | 10.1 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 1.4 | 299.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 1.2 | 7.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 1.1 | 4.3 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 1.0 | 6.9 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.8 | 1.6 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.8 | 15.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.7 | 2.0 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.7 | 2.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.5 | 1.5 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.5 | 2.6 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.5 | 3.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.5 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.5 | 9.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.4 | 2.9 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.4 | 2.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 4.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 3.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.4 | 31.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.4 | 1.1 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.4 | 3.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.3 | 1.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.3 | 4.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 1.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 1.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 2.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.3 | 5.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 2.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 1.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.3 | 0.8 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 8.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.3 | 5.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 0.8 | GO:0016590 | ACF complex(GO:0016590) |
| 0.2 | 2.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 2.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 9.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 1.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 506.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 2.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.2 | 1.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 1.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 9.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 0.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 13.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 6.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 4.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 4.0 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 37.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 2.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 3.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 1.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 6.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 1.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 2.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 2.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.8 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.7 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 5.8 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 12.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 396.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 0.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 3.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 3.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 4.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 10.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 5.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 4.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 1.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.5 | GO:0031095 | platelet dense tubular network(GO:0031094) platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 2.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 1.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.0 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 5.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 48.8 | GO:0019862 | IgA binding(GO:0019862) |
| 6.2 | 1703.1 | GO:0003823 | antigen binding(GO:0003823) |
| 2.6 | 10.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 2.1 | 8.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 1.5 | 9.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.4 | 26.5 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 1.3 | 8.9 | GO:0042806 | fucose binding(GO:0042806) |
| 1.3 | 17.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 1.1 | 3.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.1 | 4.3 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.9 | 8.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.8 | 2.5 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.8 | 5.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.8 | 7.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.7 | 2.9 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.7 | 17.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.7 | 8.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.6 | 11.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.6 | 2.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.5 | 3.2 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.5 | 2.6 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.5 | 2.6 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 1.6 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.5 | 1.6 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.5 | 4.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.5 | 2.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.5 | 2.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.5 | 1.4 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.5 | 3.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.4 | 14.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 9.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.4 | 8.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.4 | 12.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 3.8 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.4 | 1.1 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.4 | 5.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.4 | 10.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 2.8 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.3 | 10.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 1.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 7.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.3 | 1.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 1.6 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.3 | 37.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 11.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 0.9 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.3 | 0.9 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.3 | 9.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.3 | 1.8 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 3.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 2.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 1.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.3 | 0.8 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.3 | 1.4 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.3 | 2.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 2.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 2.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.3 | 1.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 7.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.4 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.2 | 3.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 1.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 1.2 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 0.9 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.2 | 2.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 1.7 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 0.6 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.2 | 1.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 1.0 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.2 | 1.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 1.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 2.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 1.0 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 1.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 0.9 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 1.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 6.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 2.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 1.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 0.8 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 4.0 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.2 | 5.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 1.6 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 1.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 0.5 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.2 | 4.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 1.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 6.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 7.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 1.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 2.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.7 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 4.6 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 5.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.8 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 1.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.4 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.1 | 2.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 6.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.7 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.4 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.9 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 4.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 3.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 2.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0050509 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 2.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 4.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.2 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.1 | 2.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.8 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.2 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 13.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 2.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 2.0 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 3.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.4 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 2.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 2.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.5 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.4 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.5 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.7 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.1 | 0.7 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 2.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.5 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 1.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 2.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.4 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 1.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 8.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 1.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 1.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.0 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.4 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 1.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 3.2 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 1.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 4.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 29.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 1.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 12.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.3 | 9.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 41.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.3 | 20.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 8.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 10.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 3.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 10.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 2.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 15.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 11.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 6.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 7.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 2.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 7.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 10.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 4.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 16.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 6.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 4.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 5.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 0.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 15.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 3.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 5.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 2.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 19.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 0.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 1.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 4.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 5.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 4.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 6.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 31.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 3.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.3 | 10.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 7.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 7.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.3 | 3.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 13.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 2.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 12.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 9.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 2.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 9.8 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.2 | 9.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 5.6 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.2 | 8.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 8.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 6.0 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 7.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.4 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 2.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 4.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 41.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 16.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 4.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 2.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 4.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 4.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 6.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 3.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 3.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 27.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 10.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 7.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 5.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.9 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 4.0 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 2.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 5.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 9.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.8 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 1.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 2.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 6.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 2.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 5.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 2.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 2.2 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |