Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for PRDM4
Z-value: 0.70
Transcription factors associated with PRDM4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM4
|
ENSG00000110851.12 | PR/SET domain 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM4 | hg38_v1_chr12_-_107760928_107760999 | 0.46 | 8.4e-03 | Click! |
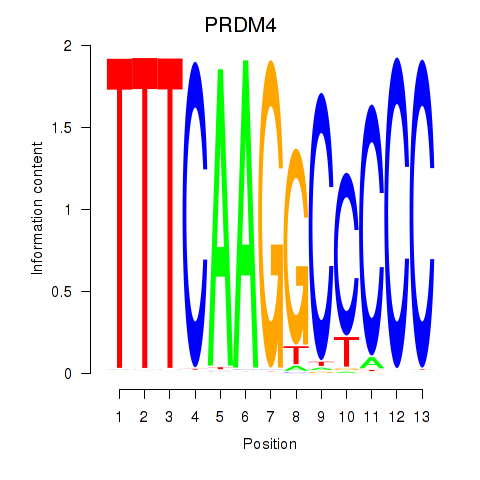
Activity profile of PRDM4 motif
Sorted Z-values of PRDM4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_40611863 | 3.03 |
ENST00000601032.5
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr7_+_91264426 | 2.59 |
ENST00000287934.4
|
FZD1
|
frizzled class receptor 1 |
| chr14_+_105486867 | 2.11 |
ENST00000409393.6
ENST00000392531.4 |
CRIP1
|
cysteine rich protein 1 |
| chr17_+_44557476 | 1.90 |
ENST00000315323.5
|
FZD2
|
frizzled class receptor 2 |
| chr1_-_42766978 | 1.67 |
ENST00000372526.2
ENST00000236040.8 ENST00000296388.10 ENST00000397054.7 |
P3H1
|
prolyl 3-hydroxylase 1 |
| chr19_-_48868590 | 1.57 |
ENST00000263265.11
|
PLEKHA4
|
pleckstrin homology domain containing A4 |
| chr2_+_218400039 | 1.52 |
ENST00000452977.5
|
CTDSP1
|
CTD small phosphatase 1 |
| chr2_+_218399838 | 1.47 |
ENST00000273062.7
|
CTDSP1
|
CTD small phosphatase 1 |
| chr19_-_48868454 | 1.43 |
ENST00000355496.9
|
PLEKHA4
|
pleckstrin homology domain containing A4 |
| chr7_-_5423826 | 1.38 |
ENST00000430969.6
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr13_+_51861963 | 1.19 |
ENST00000242819.7
|
CCDC70
|
coiled-coil domain containing 70 |
| chr17_+_7884783 | 1.17 |
ENST00000380358.9
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr22_+_38468036 | 1.14 |
ENST00000409006.3
ENST00000216014.9 |
KDELR3
|
KDEL endoplasmic reticulum protein retention receptor 3 |
| chr11_+_47257953 | 1.09 |
ENST00000437276.1
ENST00000436029.5 ENST00000467728.5 ENST00000441012.7 ENST00000405853.7 |
NR1H3
|
nuclear receptor subfamily 1 group H member 3 |
| chr7_+_143381907 | 1.09 |
ENST00000392910.6
|
ZYX
|
zyxin |
| chr1_-_68050615 | 1.08 |
ENST00000646789.1
|
DIRAS3
|
DIRAS family GTPase 3 |
| chr10_+_86968432 | 1.02 |
ENST00000416348.1
ENST00000372013.8 |
ADIRF
|
adipogenesis regulatory factor |
| chr12_+_80444228 | 1.01 |
ENST00000644991.3
|
PTPRQ
|
protein tyrosine phosphatase receptor type Q |
| chr7_+_143381561 | 0.95 |
ENST00000354434.8
|
ZYX
|
zyxin |
| chr12_-_57846686 | 0.92 |
ENST00000548823.1
ENST00000398073.7 |
CTDSP2
|
CTD small phosphatase 2 |
| chr10_-_103452384 | 0.85 |
ENST00000369788.7
|
CALHM2
|
calcium homeostasis modulator family member 2 |
| chr1_+_172533104 | 0.84 |
ENST00000616058.4
ENST00000263688.4 ENST00000610051.5 |
SUCO
|
SUN domain containing ossification factor |
| chr20_-_62937936 | 0.81 |
ENST00000266070.8
ENST00000370371.8 |
DIDO1
|
death inducer-obliterator 1 |
| chr9_+_136665745 | 0.78 |
ENST00000371698.3
|
EGFL7
|
EGF like domain multiple 7 |
| chr11_-_441964 | 0.76 |
ENST00000332826.7
|
ANO9
|
anoctamin 9 |
| chr9_-_124500986 | 0.71 |
ENST00000373587.3
|
NR5A1
|
nuclear receptor subfamily 5 group A member 1 |
| chr19_-_34773184 | 0.71 |
ENST00000588760.1
ENST00000329285.13 ENST00000587354.6 |
ZNF599
|
zinc finger protein 599 |
| chr21_+_34073569 | 0.69 |
ENST00000399312.3
ENST00000381151.5 ENST00000362077.4 |
MRPS6
SLC5A3
ENSG00000272657.1
|
mitochondrial ribosomal protein S6 solute carrier family 5 member 3 novel transcript |
| chr5_-_181261078 | 0.60 |
ENST00000611618.1
|
TRIM52
|
tripartite motif containing 52 |
| chr19_+_42220283 | 0.57 |
ENST00000301215.8
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
| chr4_-_10116845 | 0.56 |
ENST00000382451.6
ENST00000382452.6 |
WDR1
|
WD repeat domain 1 |
| chr21_+_33403391 | 0.56 |
ENST00000290219.11
ENST00000381995.5 |
IFNGR2
|
interferon gamma receptor 2 |
| chr12_-_10723307 | 0.51 |
ENST00000279550.11
ENST00000228251.9 |
YBX3
|
Y-box binding protein 3 |
| chr2_+_203706475 | 0.51 |
ENST00000374481.7
ENST00000458610.6 |
CD28
|
CD28 molecule |
| chr1_+_26787926 | 0.50 |
ENST00000674202.1
ENST00000674222.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis class V |
| chr1_+_6451578 | 0.49 |
ENST00000434576.2
ENST00000477679.2 |
ESPN
|
espin |
| chr11_+_19117123 | 0.48 |
ENST00000399351.7
ENST00000446113.7 |
ZDHHC13
|
zinc finger DHHC-type palmitoyltransferase 13 |
| chr16_-_67936808 | 0.48 |
ENST00000358514.9
|
PSMB10
|
proteasome 20S subunit beta 10 |
| chr14_-_95516616 | 0.42 |
ENST00000682763.1
|
SYNE3
|
spectrin repeat containing nuclear envelope family member 3 |
| chr8_+_93700667 | 0.42 |
ENST00000523453.5
ENST00000520955.5 |
CIBAR1
|
CBY1 interacting BAR domain containing 1 |
| chr1_+_26788166 | 0.41 |
ENST00000374145.6
ENST00000431541.6 ENST00000674273.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis class V |
| chr20_-_51023081 | 0.41 |
ENST00000433903.5
ENST00000424171.5 ENST00000371571.5 ENST00000439216.5 |
KCNG1
|
potassium voltage-gated channel modifier subfamily G member 1 |
| chr6_-_32192845 | 0.41 |
ENST00000487761.5
|
GPSM3
|
G protein signaling modulator 3 |
| chr1_-_224433776 | 0.40 |
ENST00000678879.1
ENST00000651911.2 |
WDR26
|
WD repeat domain 26 |
| chr9_+_93576557 | 0.38 |
ENST00000359246.9
ENST00000375376.8 |
PHF2
|
PHD finger protein 2 |
| chr4_-_10116779 | 0.37 |
ENST00000499869.7
|
WDR1
|
WD repeat domain 1 |
| chr8_+_93700524 | 0.37 |
ENST00000522324.5
ENST00000518322.6 ENST00000522803.5 ENST00000423990.6 ENST00000620645.1 |
CIBAR1
|
CBY1 interacting BAR domain containing 1 |
| chr15_+_66505289 | 0.35 |
ENST00000565627.5
ENST00000564179.5 ENST00000307897.10 |
ZWILCH
|
zwilch kinetochore protein |
| chr15_+_66504959 | 0.34 |
ENST00000535141.6
ENST00000613446.4 ENST00000446801.6 |
ZWILCH
|
zwilch kinetochore protein |
| chr15_-_66504832 | 0.34 |
ENST00000569438.2
ENST00000569696.5 ENST00000307961.11 |
RPL4
|
ribosomal protein L4 |
| chr6_-_32192630 | 0.32 |
ENST00000375040.8
|
GPSM3
|
G protein signaling modulator 3 |
| chr1_+_222737283 | 0.31 |
ENST00000360827.6
|
FAM177B
|
family with sequence similarity 177 member B |
| chr4_-_82373946 | 0.31 |
ENST00000352301.8
ENST00000509107.1 ENST00000353341.8 ENST00000313899.12 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D |
| chr16_+_14974974 | 0.30 |
ENST00000535621.6
ENST00000396410.9 ENST00000566426.1 |
PDXDC1
|
pyridoxal dependent decarboxylase domain containing 1 |
| chr20_+_31968141 | 0.29 |
ENST00000562532.3
|
XKR7
|
XK related 7 |
| chr9_+_17135017 | 0.28 |
ENST00000380641.4
ENST00000380647.8 |
CNTLN
|
centlein |
| chr2_+_233388146 | 0.27 |
ENST00000409813.7
|
DGKD
|
diacylglycerol kinase delta |
| chr8_-_134639808 | 0.27 |
ENST00000522257.5
|
ZFAT
|
zinc finger and AT-hook domain containing |
| chr1_+_26787667 | 0.26 |
ENST00000674335.1
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis class V |
| chr4_-_82374143 | 0.23 |
ENST00000507010.5
ENST00000503822.1 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D |
| chr5_+_103120314 | 0.19 |
ENST00000613674.4
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr12_+_69585666 | 0.19 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1 subunit 2 |
| chr16_+_14975098 | 0.18 |
ENST00000569715.5
ENST00000627450.2 |
PDXDC1
|
pyridoxal dependent decarboxylase domain containing 1 |
| chr16_+_14975061 | 0.18 |
ENST00000455313.6
|
PDXDC1
|
pyridoxal dependent decarboxylase domain containing 1 |
| chr14_-_105065422 | 0.18 |
ENST00000329797.8
ENST00000539291.6 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr10_+_102226293 | 0.18 |
ENST00000370005.4
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chrX_+_49235460 | 0.18 |
ENST00000376227.4
|
CCDC22
|
coiled-coil domain containing 22 |
| chrX_+_15790446 | 0.17 |
ENST00000380308.7
ENST00000307771.8 |
ZRSR2
|
zinc finger CCCH-type, RNA binding motif and serine/arginine rich 2 |
| chr16_+_14975149 | 0.17 |
ENST00000325823.11
|
PDXDC1
|
pyridoxal dependent decarboxylase domain containing 1 |
| chr12_+_104288889 | 0.13 |
ENST00000527335.5
|
TXNRD1
|
thioredoxin reductase 1 |
| chr4_+_673518 | 0.12 |
ENST00000506838.5
|
MYL5
|
myosin light chain 5 |
| chr22_-_21002081 | 0.11 |
ENST00000215742.9
ENST00000399133.2 |
THAP7
|
THAP domain containing 7 |
| chr1_+_171781635 | 0.11 |
ENST00000361735.4
ENST00000362019.7 ENST00000367737.9 |
EEF1AKNMT
|
eEF1A lysine and N-terminal methyltransferase |
| chr20_+_62938111 | 0.11 |
ENST00000266069.5
|
GID8
|
GID complex subunit 8 homolog |
| chrX_-_135296024 | 0.10 |
ENST00000370764.1
|
ZNF75D
|
zinc finger protein 75D |
| chr19_-_53132873 | 0.10 |
ENST00000601493.5
ENST00000599261.5 ENST00000597503.5 ENST00000500065.8 ENST00000594011.5 ENST00000595193.5 ENST00000595813.5 ENST00000600574.5 ENST00000596051.1 ENST00000601110.5 ENST00000243643.9 ENST00000421033.5 |
ZNF415
|
zinc finger protein 415 |
| chr19_-_11529094 | 0.06 |
ENST00000588998.5
ENST00000586149.1 |
ECSIT
|
ECSIT signaling integrator |
| chrX_+_116436599 | 0.03 |
ENST00000598581.3
|
SLC6A14
|
solute carrier family 6 member 14 |
| chr5_+_159916475 | 0.01 |
ENST00000306675.5
|
ADRA1B
|
adrenoceptor alpha 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.6 | 1.7 | GO:1901874 | negative regulation of post-translational protein modification(GO:1901874) |
| 0.5 | 2.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 | 0.9 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.2 | 1.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.2 | 0.8 | GO:2001226 | negative regulation of anion channel activity(GO:0010360) negative regulation of chloride transport(GO:2001226) |
| 0.2 | 1.9 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.1 | 0.7 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 1.0 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 0.7 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 1.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.5 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.4 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 1.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.5 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 2.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 1.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.7 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.5 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 1.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.8 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.5 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.4 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 1.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 3.9 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 3.0 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.6 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.4 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.2 | 0.9 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.2 | 0.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 2.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 1.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.9 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 2.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.3 | 1.7 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.3 | 3.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 4.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 2.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.6 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 3.0 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 3.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 4.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |