Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for PRRX1_ALX4_PHOX2A
Z-value: 0.89
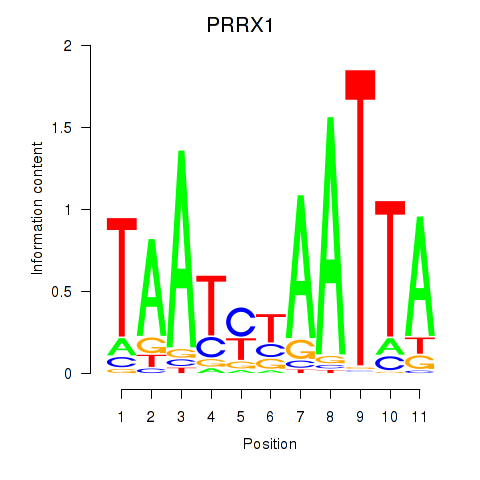
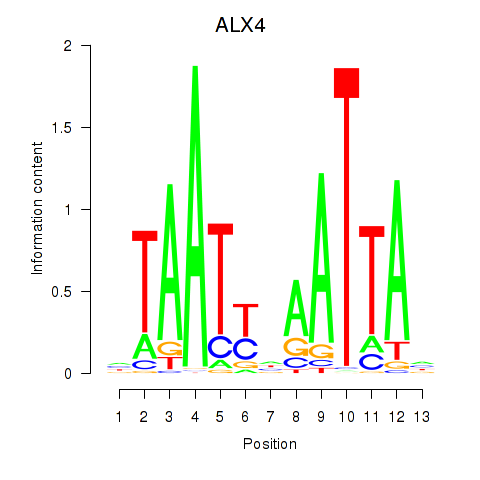
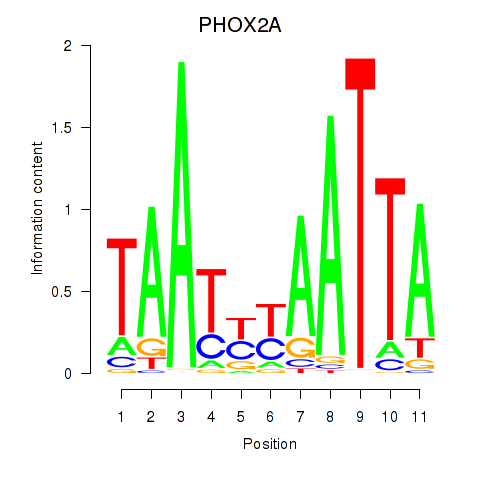
Transcription factors associated with PRRX1_ALX4_PHOX2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRRX1
|
ENSG00000116132.12 | paired related homeobox 1 |
|
ALX4
|
ENSG00000052850.8 | ALX homeobox 4 |
|
PHOX2A
|
ENSG00000165462.5 | paired like homeobox 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PHOX2A | hg38_v1_chr11_-_72244166_72244184 | -0.49 | 4.4e-03 | Click! |
| PRRX1 | hg38_v1_chr1_+_170663917_170663943 | -0.11 | 5.7e-01 | Click! |
| ALX4 | hg38_v1_chr11_-_44310126_44310152 | -0.09 | 6.4e-01 | Click! |
Activity profile of PRRX1_ALX4_PHOX2A motif
Sorted Z-values of PRRX1_ALX4_PHOX2A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_36606355 | 6.16 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr6_+_121437378 | 6.11 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr8_-_26867267 | 3.47 |
ENST00000380573.4
|
ADRA1A
|
adrenoceptor alpha 1A |
| chr3_-_195583931 | 3.46 |
ENST00000343267.8
ENST00000421243.5 ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr10_+_68106109 | 3.37 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr15_+_51377247 | 3.27 |
ENST00000396399.6
|
GLDN
|
gliomedin |
| chr6_+_54099538 | 3.17 |
ENST00000447836.6
|
MLIP
|
muscular LMNA interacting protein |
| chr6_+_54099565 | 2.93 |
ENST00000511678.5
|
MLIP
|
muscular LMNA interacting protein |
| chr15_+_51377410 | 2.90 |
ENST00000612989.1
|
GLDN
|
gliomedin |
| chr3_-_165196369 | 2.90 |
ENST00000475390.2
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr14_+_94612383 | 2.75 |
ENST00000393080.8
ENST00000555820.1 ENST00000393078.5 ENST00000467132.5 |
SERPINA3
|
serpin family A member 3 |
| chr11_+_108116688 | 2.64 |
ENST00000672284.1
|
ACAT1
|
acetyl-CoA acetyltransferase 1 |
| chr3_-_165196689 | 2.56 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr4_-_175891691 | 2.37 |
ENST00000507540.1
|
GPM6A
|
glycoprotein M6A |
| chr2_+_161416273 | 2.35 |
ENST00000389554.8
|
TBR1
|
T-box brain transcription factor 1 |
| chr4_+_154563003 | 2.30 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr15_+_51377315 | 2.25 |
ENST00000558426.5
|
GLDN
|
gliomedin |
| chr6_-_75363003 | 1.91 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr2_+_165294031 | 1.84 |
ENST00000283256.10
|
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr17_+_55264952 | 1.80 |
ENST00000226067.10
|
HLF
|
HLF transcription factor, PAR bZIP family member |
| chr13_-_101416441 | 1.71 |
ENST00000675332.1
ENST00000676315.1 ENST00000251127.11 |
NALCN
|
sodium leak channel, non-selective |
| chr18_-_24311495 | 1.65 |
ENST00000357041.8
|
OSBPL1A
|
oxysterol binding protein like 1A |
| chr4_-_175812746 | 1.63 |
ENST00000393658.6
|
GPM6A
|
glycoprotein M6A |
| chr10_-_20897288 | 1.59 |
ENST00000377122.9
|
NEBL
|
nebulette |
| chr11_+_75100531 | 1.56 |
ENST00000531713.5
|
SLCO2B1
|
solute carrier organic anion transporter family member 2B1 |
| chr6_+_63576864 | 1.56 |
ENST00000627002.1
|
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chrX_-_15314543 | 1.50 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr7_-_83162857 | 1.50 |
ENST00000333891.14
|
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr1_-_93681829 | 1.45 |
ENST00000260502.11
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr2_-_174764436 | 1.40 |
ENST00000409323.1
ENST00000261007.9 ENST00000348749.9 ENST00000672640.1 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr9_+_121567057 | 1.39 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr12_+_12725897 | 1.39 |
ENST00000326765.10
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr18_+_23134561 | 1.37 |
ENST00000400473.6
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr5_+_141364153 | 1.37 |
ENST00000518069.2
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr16_-_75726490 | 1.32 |
ENST00000640559.2
|
CPHXL
|
cytoplasmic polyadenylated homeobox like |
| chr16_-_57946602 | 1.31 |
ENST00000564654.1
|
CNGB1
|
cyclic nucleotide gated channel subunit beta 1 |
| chr6_+_54018910 | 1.28 |
ENST00000514921.5
ENST00000274897.9 ENST00000370877.6 |
MLIP
|
muscular LMNA interacting protein |
| chr7_-_108243234 | 1.25 |
ENST00000417701.5
|
NRCAM
|
neuronal cell adhesion molecule |
| chr5_-_116536458 | 1.25 |
ENST00000510263.5
|
SEMA6A
|
semaphorin 6A |
| chr1_+_207325629 | 1.24 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr21_-_34526850 | 1.24 |
ENST00000481448.5
ENST00000381132.6 |
RCAN1
|
regulator of calcineurin 1 |
| chr2_-_201739205 | 1.23 |
ENST00000681558.1
ENST00000681495.1 |
ALS2
|
alsin Rho guanine nucleotide exchange factor ALS2 |
| chr8_-_85341705 | 1.23 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr11_-_40294089 | 1.22 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr5_-_116536428 | 1.21 |
ENST00000515009.5
|
SEMA6A
|
semaphorin 6A |
| chr1_+_177170916 | 1.18 |
ENST00000361539.5
|
BRINP2
|
BMP/retinoic acid inducible neural specific 2 |
| chr5_+_141343818 | 1.18 |
ENST00000619750.1
ENST00000253812.8 |
PCDHGA3
|
protocadherin gamma subfamily A, 3 |
| chr6_+_55327469 | 1.18 |
ENST00000340465.2
|
GFRAL
|
GDNF family receptor alpha like |
| chr5_+_141338753 | 1.17 |
ENST00000528330.2
ENST00000394576.3 |
PCDHGA2
|
protocadherin gamma subfamily A, 2 |
| chr1_+_50103903 | 1.16 |
ENST00000371827.5
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr16_-_71577082 | 1.15 |
ENST00000355962.5
|
TAT
|
tyrosine aminotransferase |
| chr18_-_55322215 | 1.13 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr3_+_160225409 | 1.11 |
ENST00000326474.5
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr13_-_46897021 | 1.09 |
ENST00000542664.4
ENST00000543956.5 |
HTR2A
|
5-hydroxytryptamine receptor 2A |
| chr3_-_12545499 | 1.06 |
ENST00000564146.4
|
MKRN2OS
|
MKRN2 opposite strand |
| chr7_-_83162899 | 1.06 |
ENST00000423517.6
|
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr14_+_20239286 | 1.04 |
ENST00000641082.1
|
OR11H4
|
olfactory receptor family 11 subfamily H member 4 |
| chr2_-_174764407 | 1.03 |
ENST00000409219.5
ENST00000409542.5 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr6_-_134317905 | 1.01 |
ENST00000461976.2
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_-_9791586 | 0.98 |
ENST00000571134.2
|
DHRS7C
|
dehydrogenase/reductase 7C |
| chrX_+_77910656 | 0.98 |
ENST00000343533.9
ENST00000341514.11 ENST00000645454.1 ENST00000642651.1 ENST00000644362.1 |
ATP7A
PGK1
|
ATPase copper transporting alpha phosphoglycerate kinase 1 |
| chr16_+_7332744 | 0.95 |
ENST00000436368.6
ENST00000311745.9 ENST00000340209.8 ENST00000620507.4 |
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr3_-_196515315 | 0.94 |
ENST00000397537.3
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr19_-_7021431 | 0.94 |
ENST00000636986.2
ENST00000637800.1 |
MBD3L2B
|
methyl-CpG binding domain protein 3 like 2B |
| chr18_-_55321986 | 0.93 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr6_-_138512493 | 0.92 |
ENST00000533765.1
|
NHSL1
|
NHS like 1 |
| chr11_+_49028823 | 0.91 |
ENST00000332682.9
|
TRIM49B
|
tripartite motif containing 49B |
| chr1_+_42380772 | 0.90 |
ENST00000431473.4
ENST00000410070.6 |
RIMKLA
|
ribosomal modification protein rimK like family member A |
| chr8_-_17895403 | 0.89 |
ENST00000381840.5
ENST00000398054.5 |
FGL1
|
fibrinogen like 1 |
| chr7_-_111392915 | 0.89 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr2_-_55269038 | 0.88 |
ENST00000417363.5
ENST00000412530.1 ENST00000366137.6 ENST00000420637.5 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr16_+_6483728 | 0.88 |
ENST00000675459.1
ENST00000551752.5 |
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr19_-_11346228 | 0.88 |
ENST00000588560.5
ENST00000592952.5 |
TMEM205
|
transmembrane protein 205 |
| chr21_-_34526815 | 0.88 |
ENST00000492600.1
|
RCAN1
|
regulator of calcineurin 1 |
| chr7_+_90709530 | 0.87 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14 |
| chr4_-_99435134 | 0.84 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_-_7503744 | 0.82 |
ENST00000396620.7
ENST00000432237.3 |
CD163
|
CD163 molecule |
| chr1_+_109213887 | 0.82 |
ENST00000234677.7
ENST00000369923.4 |
SARS1
|
seryl-tRNA synthetase 1 |
| chr11_+_89924064 | 0.81 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr2_+_28778848 | 0.81 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1 catalytic subunit beta |
| chr12_-_21775581 | 0.81 |
ENST00000537950.1
ENST00000665145.1 |
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr12_-_118359166 | 0.81 |
ENST00000542902.5
|
TAOK3
|
TAO kinase 3 |
| chr12_+_20815672 | 0.81 |
ENST00000261196.6
ENST00000381541.7 ENST00000540229.1 |
SLCO1B3
SLCO1B3-SLCO1B7
|
solute carrier organic anion transporter family member 1B3 SLCO1B3-SLCO1B7 readthrough |
| chr6_+_54018992 | 0.80 |
ENST00000509997.5
|
MLIP
|
muscular LMNA interacting protein |
| chr5_+_141364231 | 0.79 |
ENST00000611914.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr5_+_173918186 | 0.79 |
ENST00000657000.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr5_+_42548043 | 0.78 |
ENST00000618088.4
ENST00000612382.4 |
GHR
|
growth hormone receptor |
| chr16_+_6483813 | 0.78 |
ENST00000675653.1
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr5_+_161850597 | 0.78 |
ENST00000634335.1
ENST00000635880.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr8_-_85341659 | 0.75 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr4_+_94995796 | 0.75 |
ENST00000506363.5
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr6_+_3258888 | 0.74 |
ENST00000380305.4
|
PSMG4
|
proteasome assembly chaperone 4 |
| chrX_+_120604199 | 0.74 |
ENST00000371315.3
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr3_+_69936629 | 0.74 |
ENST00000394348.2
ENST00000531774.1 |
MITF
|
melanocyte inducing transcription factor |
| chr20_+_33235987 | 0.72 |
ENST00000375422.6
ENST00000375413.8 ENST00000354297.9 |
BPIFA1
|
BPI fold containing family A member 1 |
| chr7_+_80626148 | 0.71 |
ENST00000428497.5
|
CD36
|
CD36 molecule |
| chr15_+_43800586 | 0.71 |
ENST00000442995.4
ENST00000458412.2 |
HYPK
|
huntingtin interacting protein K |
| chr3_+_154121366 | 0.71 |
ENST00000465093.6
ENST00000496710.5 ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor 26 |
| chr1_+_183805105 | 0.71 |
ENST00000360851.4
|
RGL1
|
ral guanine nucleotide dissociation stimulator like 1 |
| chr12_-_46825949 | 0.71 |
ENST00000547477.5
ENST00000447411.5 ENST00000266579.9 |
SLC38A4
|
solute carrier family 38 member 4 |
| chr2_+_222860942 | 0.70 |
ENST00000392066.7
ENST00000680251.1 ENST00000679541.1 ENST00000679545.1 ENST00000680395.1 |
ACSL3
|
acyl-CoA synthetase long chain family member 3 |
| chr8_-_17895487 | 0.69 |
ENST00000427924.5
ENST00000381841.4 |
FGL1
|
fibrinogen like 1 |
| chr2_+_222861005 | 0.69 |
ENST00000680147.1
ENST00000681009.1 ENST00000679514.1 ENST00000357430.8 |
ACSL3
|
acyl-CoA synthetase long chain family member 3 |
| chr19_-_11346259 | 0.69 |
ENST00000590788.1
ENST00000354882.10 ENST00000586590.5 ENST00000589555.5 ENST00000586956.5 ENST00000593256.6 ENST00000447337.5 ENST00000591677.5 ENST00000586701.1 ENST00000589655.1 |
TMEM205
RAB3D
|
transmembrane protein 205 RAB3D, member RAS oncogene family |
| chr12_-_16608183 | 0.68 |
ENST00000354662.5
ENST00000538051.5 |
LMO3
|
LIM domain only 3 |
| chr8_-_61689768 | 0.68 |
ENST00000517847.6
ENST00000389204.8 ENST00000517661.5 ENST00000517903.5 ENST00000522603.5 ENST00000541428.5 ENST00000522349.5 ENST00000522835.5 ENST00000518306.5 |
ASPH
|
aspartate beta-hydroxylase |
| chr17_+_46624036 | 0.67 |
ENST00000575068.5
|
NSF
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr5_+_173918216 | 0.67 |
ENST00000519467.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr5_-_11589019 | 0.67 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2 |
| chr6_-_136550407 | 0.67 |
ENST00000354570.8
|
MAP7
|
microtubule associated protein 7 |
| chr7_+_23299306 | 0.67 |
ENST00000466681.2
|
MALSU1
|
mitochondrial assembly of ribosomal large subunit 1 |
| chr12_-_7503841 | 0.66 |
ENST00000359156.8
|
CD163
|
CD163 molecule |
| chr18_-_66604076 | 0.66 |
ENST00000540086.5
ENST00000580157.2 ENST00000262150.7 |
CDH19
|
cadherin 19 |
| chr5_-_88884525 | 0.66 |
ENST00000502983.5
|
MEF2C
|
myocyte enhancer factor 2C |
| chr5_+_98773651 | 0.65 |
ENST00000513185.3
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr5_+_142770367 | 0.65 |
ENST00000645722.2
ENST00000274498.9 |
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr2_+_233691607 | 0.65 |
ENST00000373424.5
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chr11_+_6921002 | 0.65 |
ENST00000317834.5
|
OR2D3
|
olfactory receptor family 2 subfamily D member 3 |
| chr4_+_159267154 | 0.65 |
ENST00000510510.5
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr2_+_178320228 | 0.65 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr5_-_11588842 | 0.65 |
ENST00000503622.5
|
CTNND2
|
catenin delta 2 |
| chrY_-_6874027 | 0.65 |
ENST00000215479.10
|
AMELY
|
amelogenin Y-linked |
| chr18_+_58196736 | 0.64 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr19_-_11346196 | 0.64 |
ENST00000586218.5
|
TMEM205
|
transmembrane protein 205 |
| chr1_-_241999091 | 0.63 |
ENST00000357246.4
|
MAP1LC3C
|
microtubule associated protein 1 light chain 3 gamma |
| chr12_-_81598360 | 0.63 |
ENST00000333447.11
ENST00000407050.8 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr10_+_70093626 | 0.62 |
ENST00000678195.1
|
MACROH2A2
|
macroH2A.2 histone |
| chr11_-_58731968 | 0.62 |
ENST00000278400.3
|
GLYAT
|
glycine-N-acyltransferase |
| chr4_+_94974984 | 0.62 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr7_-_117872209 | 0.62 |
ENST00000454375.5
|
CTTNBP2
|
cortactin binding protein 2 |
| chr12_+_55492378 | 0.62 |
ENST00000548615.1
|
OR6C68
|
olfactory receptor family 6 subfamily C member 68 |
| chr8_+_66126896 | 0.62 |
ENST00000353317.9
|
TRIM55
|
tripartite motif containing 55 |
| chr5_+_137867868 | 0.60 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr11_-_4288083 | 0.60 |
ENST00000638166.1
|
SSU72P4
|
SSU72 pseudogene 4 |
| chr11_+_55262152 | 0.60 |
ENST00000417545.5
|
TRIM48
|
tripartite motif containing 48 |
| chr10_-_45315608 | 0.60 |
ENST00000553795.6
|
OR13A1
|
olfactory receptor family 13 subfamily A member 1 |
| chr15_-_26629083 | 0.59 |
ENST00000400188.7
|
GABRB3
|
gamma-aminobutyric acid type A receptor subunit beta3 |
| chr11_-_58268260 | 0.59 |
ENST00000395079.2
|
OR10W1
|
olfactory receptor family 10 subfamily W member 1 |
| chr12_-_16608073 | 0.59 |
ENST00000441439.6
|
LMO3
|
LIM domain only 3 |
| chr3_-_9249623 | 0.58 |
ENST00000383836.8
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr6_+_3258914 | 0.58 |
ENST00000438998.7
ENST00000419065.6 ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome assembly chaperone 4 |
| chr4_+_94995919 | 0.58 |
ENST00000509540.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr16_-_75701459 | 0.58 |
ENST00000633875.4
|
DUXB
|
double homeobox B |
| chr3_+_69936583 | 0.57 |
ENST00000314557.10
ENST00000394351.9 |
MITF
|
melanocyte inducing transcription factor |
| chrX_+_11293411 | 0.56 |
ENST00000348912.4
ENST00000380714.7 ENST00000380712.7 |
AMELX
|
amelogenin X-linked |
| chr2_+_75646775 | 0.56 |
ENST00000393909.7
ENST00000358788.10 ENST00000409374.5 |
MRPL19
|
mitochondrial ribosomal protein L19 |
| chr4_+_75724569 | 0.56 |
ENST00000514213.7
ENST00000264904.8 |
USO1
|
USO1 vesicle transport factor |
| chr18_-_55322334 | 0.56 |
ENST00000630720.3
|
TCF4
|
transcription factor 4 |
| chr6_-_27867581 | 0.55 |
ENST00000331442.5
|
H1-5
|
H1.5 linker histone, cluster member |
| chr12_-_81598332 | 0.55 |
ENST00000443686.7
|
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr2_-_172102893 | 0.54 |
ENST00000234198.9
ENST00000466293.2 |
DLX2
|
distal-less homeobox 2 |
| chr6_+_30914360 | 0.54 |
ENST00000421263.1
|
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr12_-_10807286 | 0.54 |
ENST00000240615.3
|
TAS2R8
|
taste 2 receptor member 8 |
| chr11_-_58731936 | 0.54 |
ENST00000344743.8
|
GLYAT
|
glycine-N-acyltransferase |
| chr7_+_90709231 | 0.53 |
ENST00000446790.5
ENST00000265741.7 |
CDK14
|
cyclin dependent kinase 14 |
| chr1_+_12857086 | 0.53 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr9_+_27524285 | 0.53 |
ENST00000276943.3
|
IFNK
|
interferon kappa |
| chr5_+_151020438 | 0.52 |
ENST00000622181.4
ENST00000614343.4 ENST00000388825.9 ENST00000521650.5 ENST00000517973.1 |
GPX3
|
glutathione peroxidase 3 |
| chr21_-_41926680 | 0.51 |
ENST00000329623.11
|
C2CD2
|
C2 calcium dependent domain containing 2 |
| chr1_+_186296267 | 0.51 |
ENST00000533951.5
ENST00000367482.8 ENST00000635041.1 ENST00000367483.8 ENST00000367485.4 ENST00000445192.7 |
PRG4
|
proteoglycan 4 |
| chr16_+_7332839 | 0.51 |
ENST00000355637.9
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr4_+_159267737 | 0.50 |
ENST00000264431.8
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr14_-_81427390 | 0.50 |
ENST00000555447.5
|
STON2
|
stonin 2 |
| chr5_-_11588796 | 0.50 |
ENST00000513598.5
|
CTNND2
|
catenin delta 2 |
| chr7_+_123601815 | 0.50 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr4_-_99435396 | 0.49 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_-_169250825 | 0.48 |
ENST00000676869.1
ENST00000676760.1 |
THBS2
|
thrombospondin 2 |
| chr2_+_186694007 | 0.48 |
ENST00000304698.10
|
FAM171B
|
family with sequence similarity 171 member B |
| chr4_-_99435336 | 0.47 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr4_+_168654724 | 0.47 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr16_+_6483379 | 0.46 |
ENST00000552089.5
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr4_-_46388713 | 0.46 |
ENST00000507069.5
|
GABRA2
|
gamma-aminobutyric acid type A receptor subunit alpha2 |
| chr4_-_71784046 | 0.46 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr8_-_10655137 | 0.46 |
ENST00000382483.4
|
RP1L1
|
RP1 like 1 |
| chr8_-_101790934 | 0.45 |
ENST00000523645.5
ENST00000520346.1 ENST00000220931.11 ENST00000522448.5 ENST00000522951.5 ENST00000522252.5 ENST00000519098.5 |
NCALD
|
neurocalcin delta |
| chr1_-_157099798 | 0.45 |
ENST00000454449.3
|
ETV3L
|
ETS variant transcription factor 3 like |
| chr11_+_24496988 | 0.45 |
ENST00000336930.11
|
LUZP2
|
leucine zipper protein 2 |
| chr9_+_74615582 | 0.45 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B |
| chr4_-_89029881 | 0.44 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13 member A |
| chr7_-_93890744 | 0.44 |
ENST00000650573.1
ENST00000222543.11 ENST00000649913.1 ENST00000647793.1 |
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr9_-_92536645 | 0.44 |
ENST00000395534.2
|
ECM2
|
extracellular matrix protein 2 |
| chr7_+_123601836 | 0.44 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr16_-_21864103 | 0.44 |
ENST00000541674.5
|
NPIPB4
|
nuclear pore complex interacting protein family member B4 |
| chr5_+_141402764 | 0.43 |
ENST00000573521.2
ENST00000616887.1 |
PCDHGA9
|
protocadherin gamma subfamily A, 9 |
| chr3_-_87276577 | 0.43 |
ENST00000344265.8
ENST00000350375.7 |
POU1F1
|
POU class 1 homeobox 1 |
| chr7_+_138460238 | 0.42 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr1_+_248095184 | 0.42 |
ENST00000358120.3
ENST00000641893.1 |
OR2L13
|
olfactory receptor family 2 subfamily L member 13 |
| chr4_+_87832917 | 0.42 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr1_+_12916610 | 0.42 |
ENST00000616979.4
|
PRAMEF7
|
PRAME family member 7 |
| chr12_-_10826358 | 0.42 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chr3_+_35642159 | 0.41 |
ENST00000187397.8
|
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr11_-_82997394 | 0.41 |
ENST00000525117.5
ENST00000532548.5 |
RAB30
|
RAB30, member RAS oncogene family |
| chr3_-_33718207 | 0.41 |
ENST00000359576.9
ENST00000682230.1 ENST00000399362.8 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr11_+_113908983 | 0.41 |
ENST00000537778.5
|
HTR3B
|
5-hydroxytryptamine receptor 3B |
| chr2_-_182427014 | 0.40 |
ENST00000409365.5
ENST00000351439.9 |
PDE1A
|
phosphodiesterase 1A |
| chr6_+_30163541 | 0.39 |
ENST00000376694.9
|
TRIM15
|
tripartite motif containing 15 |
| chr7_+_123601859 | 0.39 |
ENST00000437535.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr4_+_87650277 | 0.39 |
ENST00000339673.11
ENST00000282479.8 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr12_-_75390890 | 0.39 |
ENST00000552497.5
ENST00000551829.5 ENST00000436898.5 |
CAPS2
|
calcyphosine 2 |
| chr10_+_24466487 | 0.38 |
ENST00000396446.5
ENST00000396445.5 ENST00000376451.4 |
KIAA1217
|
KIAA1217 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRRX1_ALX4_PHOX2A
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 1.2 | 3.5 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.9 | 3.5 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.9 | 2.6 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.7 | 2.6 | GO:0006550 | isoleucine catabolic process(GO:0006550) acetyl-CoA catabolic process(GO:0046356) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.5 | 6.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.5 | 1.8 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.4 | 1.8 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.4 | 9.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 | 1.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.3 | 2.3 | GO:0021764 | amygdala development(GO:0021764) |
| 0.3 | 1.0 | GO:0060003 | copper ion export(GO:0060003) |
| 0.3 | 0.9 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.3 | 2.4 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.2 | 1.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.2 | 2.0 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.2 | 0.7 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.2 | 0.9 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.2 | 0.9 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 1.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 1.2 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.2 | 0.7 | GO:0044407 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.2 | 1.4 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.2 | 0.7 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 0.7 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.2 | 0.7 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 1.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.2 | 2.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.5 | GO:0061075 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 3.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 1.7 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.4 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.1 | 0.4 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.1 | 1.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 1.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.2 | GO:0021622 | oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 0.3 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.8 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.1 | 1.0 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 1.8 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 2.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 1.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.1 | 0.9 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.2 | GO:0034758 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.4 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.6 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.5 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 1.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.6 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 0.5 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.7 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.7 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.1 | 0.2 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 0.7 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 2.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 1.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 2.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 4.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 1.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 1.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 4.1 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 6.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 2.4 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 1.2 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 0.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 3.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.9 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 2.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 1.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 7.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 1.3 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 8.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.3 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.4 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.3 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 1.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.4 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.2 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 1.0 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 1.2 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.5 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.4 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.2 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.8 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.3 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0099569 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.3 | 1.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.3 | 3.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 0.8 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 1.4 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.2 | 0.9 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 1.5 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 6.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.3 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.8 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 4.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.5 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 2.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.8 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 8.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 1.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 1.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 6.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 6.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 4.3 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.0 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 10.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 2.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.7 | 6.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.7 | 2.6 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.5 | 1.8 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.4 | 1.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.4 | 6.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 1.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.4 | 9.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.3 | 1.0 | GO:0004618 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 2.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 2.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 0.6 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.2 | 0.9 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.2 | 1.3 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 0.8 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.2 | 1.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 0.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 2.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.8 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 1.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 2.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.5 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.7 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 2.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.8 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.7 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 0.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 3.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 1.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.4 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 1.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 2.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 1.0 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 1.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 0.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 3.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.3 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.1 | 1.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.7 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 2.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 1.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.6 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 1.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 2.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 4.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 2.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 4.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 5.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 2.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 2.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 10.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 3.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 4.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 6.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 3.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 2.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 2.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 4.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |