|
chr14_-_52950992
Show fit
|
9.49 |
ENST00000343279.8
ENST00000399304.7
ENST00000395631.6
ENST00000341590.8
|
FERMT2
|
fermitin family member 2
|
|
chr8_-_79767462
Show fit
|
8.48 |
ENST00000674295.1
ENST00000518733.1
ENST00000674418.1
ENST00000674358.1
ENST00000354724.8
|
HEY1
|
hes related family bHLH transcription factor with YRPW motif 1
|
|
chr3_-_100993507
Show fit
|
6.68 |
ENST00000284322.10
|
ABI3BP
|
ABI family member 3 binding protein
|
|
chr3_-_100993409
Show fit
|
6.17 |
ENST00000471714.6
|
ABI3BP
|
ABI family member 3 binding protein
|
|
chr3_-_100993448
Show fit
|
6.17 |
ENST00000495063.6
ENST00000486770.7
ENST00000530539.2
|
ABI3BP
|
ABI family member 3 binding protein
|
|
chr3_+_142723999
Show fit
|
6.13 |
ENST00000476941.6
ENST00000273482.10
|
TRPC1
|
transient receptor potential cation channel subfamily C member 1
|
|
chr12_+_12785652
Show fit
|
5.30 |
ENST00000356591.5
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr11_+_118607598
Show fit
|
5.02 |
ENST00000600882.6
ENST00000356063.9
|
PHLDB1
|
pleckstrin homology like domain family B member 1
|
|
chr20_+_31637905
Show fit
|
4.93 |
ENST00000376075.4
|
COX4I2
|
cytochrome c oxidase subunit 4I2
|
|
chr6_+_125749623
Show fit
|
4.75 |
ENST00000368364.4
|
HEY2
|
hes related family bHLH transcription factor with YRPW motif 2
|
|
chr20_+_44582549
Show fit
|
4.09 |
ENST00000372886.6
|
PKIG
|
cAMP-dependent protein kinase inhibitor gamma
|
|
chr7_-_120858066
Show fit
|
4.01 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12
|
|
chr7_+_94394886
Show fit
|
4.00 |
ENST00000297268.11
ENST00000620463.1
|
COL1A2
|
collagen type I alpha 2 chain
|
|
chr9_+_70043840
Show fit
|
3.98 |
ENST00000377182.5
|
MAMDC2
|
MAM domain containing 2
|
|
chr6_-_93419545
Show fit
|
3.84 |
ENST00000369297.1
ENST00000369303.9
ENST00000680224.1
ENST00000681532.1
ENST00000679565.1
|
EPHA7
|
EPH receptor A7
|
|
chr17_-_41624541
Show fit
|
3.79 |
ENST00000540235.5
ENST00000311208.13
|
KRT17
|
keratin 17
|
|
chr11_+_112961247
Show fit
|
3.64 |
ENST00000621518.4
ENST00000618266.4
ENST00000615112.4
ENST00000615285.4
ENST00000619839.4
ENST00000401611.6
ENST00000621128.4
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr8_+_67952028
Show fit
|
3.56 |
ENST00000288368.5
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 2
|
|
chr10_-_95441015
Show fit
|
3.38 |
ENST00000371241.5
ENST00000354106.7
ENST00000371239.5
ENST00000361941.7
ENST00000277982.9
ENST00000371245.7
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr9_+_99821846
Show fit
|
3.30 |
ENST00000338488.8
ENST00000618101.4
|
NR4A3
|
nuclear receptor subfamily 4 group A member 3
|
|
chr12_+_65278919
Show fit
|
3.30 |
ENST00000538045.5
ENST00000642411.1
ENST00000535239.5
ENST00000614640.4
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr12_+_65279092
Show fit
|
3.23 |
ENST00000646299.1
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr12_+_65279445
Show fit
|
3.20 |
ENST00000642404.1
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr8_-_22232020
Show fit
|
3.19 |
ENST00000454243.7
ENST00000321613.7
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr6_+_30884353
Show fit
|
3.07 |
ENST00000428153.6
ENST00000376568.8
ENST00000452441.5
ENST00000515219.5
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr6_-_75493629
Show fit
|
3.04 |
ENST00000393004.6
|
FILIP1
|
filamin A interacting protein 1
|
|
chr17_-_58529344
Show fit
|
3.04 |
ENST00000317268.7
|
SEPTIN4
|
septin 4
|
|
chr9_+_99821876
Show fit
|
3.01 |
ENST00000395097.7
|
NR4A3
|
nuclear receptor subfamily 4 group A member 3
|
|
chr11_+_112961402
Show fit
|
3.00 |
ENST00000613217.4
ENST00000316851.12
ENST00000620046.4
ENST00000531044.5
ENST00000529356.5
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr8_-_79767843
Show fit
|
3.00 |
ENST00000337919.9
|
HEY1
|
hes related family bHLH transcription factor with YRPW motif 1
|
|
chr6_-_75493773
Show fit
|
2.99 |
ENST00000237172.12
|
FILIP1
|
filamin A interacting protein 1
|
|
chr6_+_30884063
Show fit
|
2.90 |
ENST00000511510.5
ENST00000376569.7
ENST00000376570.8
ENST00000504927.5
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr17_-_58529303
Show fit
|
2.83 |
ENST00000580844.5
|
SEPTIN4
|
septin 4
|
|
chr10_-_37857582
Show fit
|
2.82 |
ENST00000395867.8
ENST00000611278.4
|
ZNF248
|
zinc finger protein 248
|
|
chr1_-_39639626
Show fit
|
2.79 |
ENST00000372852.4
|
HEYL
|
hes related family bHLH transcription factor with YRPW motif like
|
|
chr12_+_6200759
Show fit
|
2.74 |
ENST00000645565.1
ENST00000382515.7
|
CD9
|
CD9 molecule
|
|
chr17_-_58529277
Show fit
|
2.56 |
ENST00000579371.5
|
SEPTIN4
|
septin 4
|
|
chr12_+_6828377
Show fit
|
2.50 |
ENST00000290510.10
|
P3H3
|
prolyl 3-hydroxylase 3
|
|
chr12_-_16606795
Show fit
|
2.50 |
ENST00000447609.5
|
LMO3
|
LIM domain only 3
|
|
chr11_+_111541326
Show fit
|
2.47 |
ENST00000530962.5
ENST00000528924.4
|
LAYN
|
layilin
|
|
chr3_-_45915698
Show fit
|
2.45 |
ENST00000539217.5
|
LZTFL1
|
leucine zipper transcription factor like 1
|
|
chr11_-_119340816
Show fit
|
2.45 |
ENST00000528368.3
|
C1QTNF5
|
C1q and TNF related 5
|
|
chr19_+_3094348
Show fit
|
2.36 |
ENST00000078429.9
|
GNA11
|
G protein subunit alpha 11
|
|
chr19_+_50203607
Show fit
|
2.35 |
ENST00000642316.2
ENST00000425460.6
ENST00000440075.6
ENST00000376970.6
ENST00000599920.5
|
MYH14
|
myosin heavy chain 14
|
|
chr11_+_112961480
Show fit
|
2.35 |
ENST00000621850.4
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr4_-_86594037
Show fit
|
2.32 |
ENST00000641050.1
ENST00000641831.1
ENST00000515400.3
ENST00000641391.1
ENST00000641157.1
ENST00000641737.1
ENST00000502302.6
ENST00000640527.1
ENST00000512046.2
ENST00000513186.7
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr2_+_48568981
Show fit
|
2.32 |
ENST00000394754.5
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough
|
|
chr4_-_185657520
Show fit
|
2.31 |
ENST00000438278.5
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_+_159852933
Show fit
|
2.31 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr16_-_70685975
Show fit
|
2.31 |
ENST00000338779.11
|
MTSS2
|
MTSS I-BAR domain containing 2
|
|
chr2_+_43774033
Show fit
|
2.29 |
ENST00000260605.12
ENST00000406852.7
ENST00000398823.6
ENST00000605786.5
|
DYNC2LI1
|
dynein cytoplasmic 2 light intermediate chain 1
|
|
chr4_+_155667198
Show fit
|
2.28 |
ENST00000296518.11
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1
|
|
chr7_-_16465728
Show fit
|
2.24 |
ENST00000307068.5
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr7_-_120858303
Show fit
|
2.12 |
ENST00000415871.5
ENST00000430985.1
|
TSPAN12
|
tetraspanin 12
|
|
chr16_-_70685791
Show fit
|
2.08 |
ENST00000616026.4
|
MTSS2
|
MTSS I-BAR domain containing 2
|
|
chr1_-_84893166
Show fit
|
2.06 |
ENST00000370611.4
|
LPAR3
|
lysophosphatidic acid receptor 3
|
|
chr16_-_57797764
Show fit
|
2.02 |
ENST00000465878.6
ENST00000561524.5
|
KIFC3
|
kinesin family member C3
|
|
chr4_+_155667096
Show fit
|
1.99 |
ENST00000393832.7
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1
|
|
chr11_+_118607579
Show fit
|
1.98 |
ENST00000530708.4
|
PHLDB1
|
pleckstrin homology like domain family B member 1
|
|
chr10_+_31319125
Show fit
|
1.93 |
ENST00000320985.14
ENST00000560721.6
ENST00000558440.5
ENST00000424869.6
ENST00000542815.7
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr14_+_51489112
Show fit
|
1.90 |
ENST00000356218.8
|
FRMD6
|
FERM domain containing 6
|
|
chr10_-_33334898
Show fit
|
1.87 |
ENST00000395995.5
|
NRP1
|
neuropilin 1
|
|
chr10_-_33334625
Show fit
|
1.86 |
ENST00000374875.5
ENST00000374822.8
ENST00000374867.7
|
NRP1
|
neuropilin 1
|
|
chr14_-_92748570
Show fit
|
1.85 |
ENST00000553918.1
ENST00000555699.5
ENST00000334869.9
ENST00000553802.5
ENST00000554397.5
ENST00000554919.5
ENST00000554080.5
ENST00000553371.1
ENST00000557434.5
ENST00000393218.6
|
LGMN
|
legumain
|
|
chr7_-_22220226
Show fit
|
1.85 |
ENST00000420196.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5
|
|
chr10_+_122560639
Show fit
|
1.85 |
ENST00000344338.7
ENST00000330163.8
ENST00000652446.2
ENST00000666315.1
ENST00000368955.7
ENST00000368909.7
ENST00000368956.6
ENST00000619379.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr16_+_23302292
Show fit
|
1.83 |
ENST00000343070.7
|
SCNN1B
|
sodium channel epithelial 1 subunit beta
|
|
chr17_-_28726186
Show fit
|
1.81 |
ENST00000292090.8
|
TLCD1
|
TLC domain containing 1
|
|
chr12_-_16607087
Show fit
|
1.80 |
ENST00000540445.5
|
LMO3
|
LIM domain only 3
|
|
chr15_+_57599411
Show fit
|
1.80 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein
|
|
chr10_-_33334382
Show fit
|
1.78 |
ENST00000374823.9
ENST00000374821.9
ENST00000374816.7
|
NRP1
|
neuropilin 1
|
|
chr10_-_107164692
Show fit
|
1.76 |
ENST00000263054.11
|
SORCS1
|
sortilin related VPS10 domain containing receptor 1
|
|
chr3_+_194136138
Show fit
|
1.74 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1
|
|
chr3_-_19934189
Show fit
|
1.74 |
ENST00000295824.14
|
EFHB
|
EF-hand domain family member B
|
|
chr11_-_119340544
Show fit
|
1.71 |
ENST00000530681.2
|
C1QTNF5
|
C1q and TNF related 5
|
|
chr16_-_1611985
Show fit
|
1.71 |
ENST00000426508.7
|
IFT140
|
intraflagellar transport 140
|
|
chr3_+_156142962
Show fit
|
1.70 |
ENST00000471742.5
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1
|
|
chr10_+_111077021
Show fit
|
1.69 |
ENST00000280155.4
|
ADRA2A
|
adrenoceptor alpha 2A
|
|
chr2_+_149330506
Show fit
|
1.69 |
ENST00000334166.9
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr10_-_37858037
Show fit
|
1.68 |
ENST00000395873.7
ENST00000357328.8
ENST00000395874.2
|
ZNF248
|
zinc finger protein 248
|
|
chr15_+_59438149
Show fit
|
1.61 |
ENST00000288228.10
ENST00000559628.5
ENST00000557914.5
ENST00000560474.5
|
FAM81A
|
family with sequence similarity 81 member A
|
|
chr18_+_68715191
Show fit
|
1.59 |
ENST00000578970.5
ENST00000582371.5
ENST00000584775.5
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr4_+_155667654
Show fit
|
1.58 |
ENST00000513574.1
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1
|
|
chr5_-_116536458
Show fit
|
1.55 |
ENST00000510263.5
|
SEMA6A
|
semaphorin 6A
|
|
chr12_-_48865863
Show fit
|
1.52 |
ENST00000309739.6
|
RND1
|
Rho family GTPase 1
|
|
chr1_+_61404076
Show fit
|
1.49 |
ENST00000357977.5
|
NFIA
|
nuclear factor I A
|
|
chr5_-_140564245
Show fit
|
1.49 |
ENST00000412920.7
ENST00000511201.2
ENST00000354402.9
ENST00000356738.6
|
APBB3
|
amyloid beta precursor protein binding family B member 3
|
|
chr16_-_57798008
Show fit
|
1.46 |
ENST00000421376.6
|
KIFC3
|
kinesin family member C3
|
|
chr5_+_38445539
Show fit
|
1.45 |
ENST00000397210.7
ENST00000506135.5
ENST00000508131.5
|
EGFLAM
|
EGF like, fibronectin type III and laminin G domains
|
|
chr1_-_85404494
Show fit
|
1.44 |
ENST00000633113.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr9_-_137028878
Show fit
|
1.41 |
ENST00000625103.1
ENST00000614293.4
|
ABCA2
|
ATP binding cassette subfamily A member 2
|
|
chr5_+_178895892
Show fit
|
1.38 |
ENST00000520660.5
ENST00000361362.7
ENST00000520805.5
|
ZFP2
|
ZFP2 zinc finger protein
|
|
chr4_+_183905266
Show fit
|
1.33 |
ENST00000308497.9
|
STOX2
|
storkhead box 2
|
|
chr10_+_100999287
Show fit
|
1.33 |
ENST00000370220.1
|
LZTS2
|
leucine zipper tumor suppressor 2
|
|
chr10_-_33335074
Show fit
|
1.32 |
ENST00000432372.6
|
NRP1
|
neuropilin 1
|
|
chr2_+_169733811
Show fit
|
1.31 |
ENST00000392647.7
|
KLHL23
|
kelch like family member 23
|
|
chrX_+_106802660
Show fit
|
1.26 |
ENST00000357242.10
ENST00000310452.6
ENST00000481617.6
ENST00000276175.7
|
TBC1D8B
|
TBC1 domain family member 8B
|
|
chr17_-_41624803
Show fit
|
1.25 |
ENST00000463128.5
|
KRT17
|
keratin 17
|
|
chr6_-_134052594
Show fit
|
1.25 |
ENST00000275230.6
|
SLC2A12
|
solute carrier family 2 member 12
|
|
chr16_-_10182754
Show fit
|
1.21 |
ENST00000396573.6
ENST00000675398.1
|
GRIN2A
|
glutamate ionotropic receptor NMDA type subunit 2A
|
|
chr5_+_147878703
Show fit
|
1.19 |
ENST00000296694.5
|
SCGB3A2
|
secretoglobin family 3A member 2
|
|
chr10_+_122560751
Show fit
|
1.18 |
ENST00000338354.10
ENST00000664692.1
ENST00000653442.1
ENST00000664974.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr17_-_38735577
Show fit
|
1.17 |
ENST00000610747.1
|
PCGF2
|
polycomb group ring finger 2
|
|
chr11_-_32430811
Show fit
|
1.16 |
ENST00000379079.8
ENST00000530998.5
|
WT1
|
WT1 transcription factor
|
|
chr19_-_45792755
Show fit
|
1.14 |
ENST00000377735.7
ENST00000270223.7
|
DMWD
|
DM1 locus, WD repeat containing
|
|
chr4_-_119628007
Show fit
|
1.12 |
ENST00000420633.1
ENST00000394439.5
|
PDE5A
|
phosphodiesterase 5A
|
|
chr1_-_1000139
Show fit
|
1.11 |
ENST00000428771.6
|
HES4
|
hes family bHLH transcription factor 4
|
|
chr10_+_113679523
Show fit
|
1.09 |
ENST00000345633.8
ENST00000614447.4
ENST00000369321.6
|
CASP7
|
caspase 7
|
|
chr2_+_218959635
Show fit
|
1.09 |
ENST00000302625.6
|
CDK5R2
|
cyclin dependent kinase 5 regulatory subunit 2
|
|
chr7_+_128862841
Show fit
|
1.07 |
ENST00000249289.5
ENST00000492758.1
|
ATP6V1F
|
ATPase H+ transporting V1 subunit F
|
|
chr10_+_113679839
Show fit
|
1.07 |
ENST00000369318.8
ENST00000369315.5
|
CASP7
|
caspase 7
|
|
chr3_-_100840109
Show fit
|
1.06 |
ENST00000533795.5
|
ABI3BP
|
ABI family member 3 binding protein
|
|
chr8_-_100336184
Show fit
|
1.05 |
ENST00000519527.5
ENST00000522369.5
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase
|
|
chr4_-_175812236
Show fit
|
1.03 |
ENST00000505375.5
|
GPM6A
|
glycoprotein M6A
|
|
chr16_+_53703963
Show fit
|
1.00 |
ENST00000636218.1
ENST00000637001.1
ENST00000471389.6
ENST00000637969.1
ENST00000640179.1
|
FTO
|
FTO alpha-ketoglutarate dependent dioxygenase
|
|
chr20_+_31475278
Show fit
|
1.00 |
ENST00000201979.3
|
REM1
|
RRAD and GEM like GTPase 1
|
|
chr11_+_66744618
Show fit
|
1.00 |
ENST00000524551.5
ENST00000525908.6
ENST00000540737.7
ENST00000527634.5
|
C11orf80
|
chromosome 11 open reading frame 80
|
|
chr16_-_57798091
Show fit
|
0.99 |
ENST00000562503.5
|
KIFC3
|
kinesin family member C3
|
|
chr4_-_175812746
Show fit
|
0.99 |
ENST00000393658.6
|
GPM6A
|
glycoprotein M6A
|
|
chr7_-_120857124
Show fit
|
0.98 |
ENST00000441017.5
ENST00000424710.5
ENST00000433758.5
|
TSPAN12
|
tetraspanin 12
|
|
chr15_-_43879835
Show fit
|
0.95 |
ENST00000636859.1
|
FRMD5
|
FERM domain containing 5
|
|
chr1_+_43389874
Show fit
|
0.95 |
ENST00000372450.8
|
SZT2
|
SZT2 subunit of KICSTOR complex
|
|
chr1_-_1000088
Show fit
|
0.95 |
ENST00000304952.11
|
HES4
|
hes family bHLH transcription factor 4
|
|
chr19_-_40218339
Show fit
|
0.93 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B
|
|
chr7_+_30771388
Show fit
|
0.93 |
ENST00000265299.6
|
MINDY4
|
MINDY lysine 48 deubiquitinase 4
|
|
chr4_+_26319636
Show fit
|
0.92 |
ENST00000342295.6
ENST00000506956.5
ENST00000512671.6
ENST00000345843.8
|
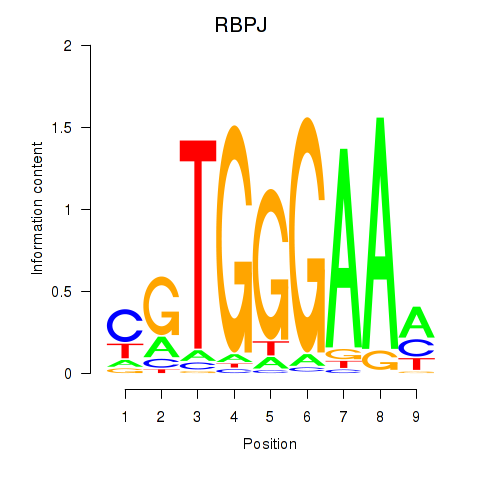
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr6_-_32224060
Show fit
|
0.89 |
ENST00000375023.3
|
NOTCH4
|
notch receptor 4
|
|
chr11_-_119340914
Show fit
|
0.89 |
ENST00000634633.1
|
C1QTNF5
|
C1q and TNF related 5
|
|
chr16_-_10182394
Show fit
|
0.88 |
ENST00000330684.4
|
GRIN2A
|
glutamate ionotropic receptor NMDA type subunit 2A
|
|
chr5_+_178941186
Show fit
|
0.88 |
ENST00000320129.7
ENST00000519564.2
|
ZNF454
|
zinc finger protein 454
|
|
chr16_-_53703810
Show fit
|
0.87 |
ENST00000569716.1
ENST00000562588.5
ENST00000621565.5
ENST00000562230.5
ENST00000563746.5
ENST00000568653.7
ENST00000647211.2
|
RPGRIP1L
|
RPGRIP1 like
|
|
chr1_+_43389889
Show fit
|
0.87 |
ENST00000562955.2
ENST00000634258.3
|
SZT2
|
SZT2 subunit of KICSTOR complex
|
|
chr5_-_88877967
Show fit
|
0.86 |
ENST00000508610.5
ENST00000636294.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr5_-_124744513
Show fit
|
0.84 |
ENST00000504926.5
|
ZNF608
|
zinc finger protein 608
|
|
chr1_+_14945775
Show fit
|
0.84 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr11_-_83682385
Show fit
|
0.83 |
ENST00000426717.6
|
DLG2
|
discs large MAGUK scaffold protein 2
|
|
chr9_+_706841
Show fit
|
0.82 |
ENST00000382293.7
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chrX_+_147911910
Show fit
|
0.82 |
ENST00000370475.9
|
FMR1
|
FMRP translational regulator 1
|
|
chr17_+_50274447
Show fit
|
0.81 |
ENST00000507382.2
|
TMEM92
|
transmembrane protein 92
|
|
chr8_-_126557691
Show fit
|
0.80 |
ENST00000652209.1
|
LRATD2
|
LRAT domain containing 2
|
|
chr2_-_40430257
Show fit
|
0.79 |
ENST00000408028.6
ENST00000332839.8
ENST00000406391.2
ENST00000405901.7
|
SLC8A1
|
solute carrier family 8 member A1
|
|
chr3_-_132037800
Show fit
|
0.79 |
ENST00000617767.4
|
CPNE4
|
copine 4
|
|
chr12_-_31792290
Show fit
|
0.77 |
ENST00000340398.5
|
H3-5
|
H3.5 histone
|
|
chr8_-_86230360
Show fit
|
0.76 |
ENST00000419776.2
ENST00000297524.8
|
SLC7A13
|
solute carrier family 7 member 13
|
|
chr5_-_116536428
Show fit
|
0.76 |
ENST00000515009.5
|
SEMA6A
|
semaphorin 6A
|
|
chr20_-_46308485
Show fit
|
0.75 |
ENST00000537909.4
|
CDH22
|
cadherin 22
|
|
chr1_-_999981
Show fit
|
0.75 |
ENST00000484667.2
|
HES4
|
hes family bHLH transcription factor 4
|
|
chr2_-_218270099
Show fit
|
0.73 |
ENST00000248450.9
ENST00000444053.5
|
AAMP
|
angio associated migratory cell protein
|
|
chr11_-_101583985
Show fit
|
0.72 |
ENST00000344327.8
|
TRPC6
|
transient receptor potential cation channel subfamily C member 6
|
|
chr2_+_37950476
Show fit
|
0.71 |
ENST00000402091.3
|
RMDN2
|
regulator of microtubule dynamics 2
|
|
chr1_-_43389768
Show fit
|
0.71 |
ENST00000372455.4
ENST00000372457.9
ENST00000290663.10
|
MED8
|
mediator complex subunit 8
|
|
chr2_+_218270392
Show fit
|
0.70 |
ENST00000248451.7
ENST00000273077.9
|
PNKD
|
PNKD metallo-beta-lactamase domain containing
|
|
chr3_-_171771295
Show fit
|
0.70 |
ENST00000418087.1
|
PLD1
|
phospholipase D1
|
|
chr12_+_106582996
Show fit
|
0.69 |
ENST00000392842.6
|
RFX4
|
regulatory factor X4
|
|
chr1_+_113390495
Show fit
|
0.69 |
ENST00000307546.14
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3
|
|
chr1_-_180502536
Show fit
|
0.66 |
ENST00000367595.4
|
ACBD6
|
acyl-CoA binding domain containing 6
|
|
chr17_+_2796404
Show fit
|
0.65 |
ENST00000366401.8
ENST00000254695.13
ENST00000542807.1
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr13_-_24922788
Show fit
|
0.65 |
ENST00000381884.9
|
CENPJ
|
centromere protein J
|
|
chr1_-_147172456
Show fit
|
0.65 |
ENST00000254101.4
|
PRKAB2
|
protein kinase AMP-activated non-catalytic subunit beta 2
|
|
chr8_+_22056966
Show fit
|
0.65 |
ENST00000517804.5
|
DMTN
|
dematin actin binding protein
|
|
chr11_+_66744831
Show fit
|
0.63 |
ENST00000642265.1
ENST00000532565.6
|
C11orf80
|
chromosome 11 open reading frame 80
|
|
chr19_+_34926892
Show fit
|
0.63 |
ENST00000303586.11
ENST00000601142.2
ENST00000439785.5
ENST00000601540.5
ENST00000601957.5
|
ZNF30
|
zinc finger protein 30
|
|
chr13_-_20230970
Show fit
|
0.63 |
ENST00000644667.1
ENST00000646108.1
|
GJB6
|
gap junction protein beta 6
|
|
chrX_+_147911943
Show fit
|
0.62 |
ENST00000621453.4
ENST00000218200.12
ENST00000370471.7
ENST00000440235.6
ENST00000370477.5
ENST00000621987.4
|
FMR1
|
FMRP translational regulator 1
|
|
chr1_+_239719542
Show fit
|
0.62 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor muscarinic 3
|
|
chr1_-_223364059
Show fit
|
0.62 |
ENST00000343846.7
ENST00000484758.6
ENST00000344029.6
ENST00000366878.9
ENST00000494793.6
ENST00000681285.1
ENST00000680429.1
ENST00000681669.1
ENST00000681305.1
|
SUSD4
|
sushi domain containing 4
|
|
chrX_-_153830527
Show fit
|
0.62 |
ENST00000393758.7
ENST00000544474.5
|
PDZD4
|
PDZ domain containing 4
|
|
chr16_-_53703883
Show fit
|
0.61 |
ENST00000262135.9
ENST00000564374.5
ENST00000566096.5
|
RPGRIP1L
|
RPGRIP1 like
|
|
chr19_+_39997031
Show fit
|
0.61 |
ENST00000599504.5
ENST00000596894.5
ENST00000601138.5
ENST00000347077.9
ENST00000600094.5
|
ZNF546
|
zinc finger protein 546
|
|
chr3_+_37975773
Show fit
|
0.59 |
ENST00000436654.1
|
CTDSPL
|
CTD small phosphatase like
|
|
chr19_-_38899800
Show fit
|
0.59 |
ENST00000414941.5
ENST00000358931.9
ENST00000392081.6
|
SIRT2
|
sirtuin 2
|
|
chr6_-_41163182
Show fit
|
0.57 |
ENST00000338469.3
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr1_-_23369813
Show fit
|
0.56 |
ENST00000314011.9
|
ZNF436
|
zinc finger protein 436
|
|
chr21_-_6499202
Show fit
|
0.56 |
ENST00000619610.2
ENST00000610664.5
ENST00000639996.1
|
U2AF1L5
|
U2 small nuclear RNA auxiliary factor 1 like 5
|
|
chr6_-_31542339
Show fit
|
0.55 |
ENST00000458640.5
|
DDX39B
|
DExD-box helicase 39B
|
|
chr8_+_106726012
Show fit
|
0.53 |
ENST00000449762.6
ENST00000297447.10
|
OXR1
|
oxidation resistance 1
|
|
chr10_+_122560679
Show fit
|
0.52 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr16_-_57802401
Show fit
|
0.51 |
ENST00000569112.5
ENST00000445690.7
ENST00000562311.5
ENST00000379655.8
|
KIFC3
|
kinesin family member C3
|
|
chr11_+_64285839
Show fit
|
0.50 |
ENST00000438980.7
ENST00000540370.1
|
GPR137
|
G protein-coupled receptor 137
|
|
chr17_-_81128873
Show fit
|
0.49 |
ENST00000572339.5
|
AATK
|
apoptosis associated tyrosine kinase
|
|
chr5_-_138178599
Show fit
|
0.49 |
ENST00000454473.5
ENST00000418329.5
ENST00000254900.10
ENST00000230901.9
ENST00000402931.5
ENST00000411594.6
ENST00000430331.1
|
BRD8
|
bromodomain containing 8
|
|
chr4_-_119627631
Show fit
|
0.48 |
ENST00000264805.9
|
PDE5A
|
phosphodiesterase 5A
|
|
chr11_+_64285219
Show fit
|
0.45 |
ENST00000377702.8
|
GPR137
|
G protein-coupled receptor 137
|
|
chr18_-_72638510
Show fit
|
0.45 |
ENST00000581073.1
|
CBLN2
|
cerebellin 2 precursor
|
|
chr15_+_67125707
Show fit
|
0.44 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3
|
|
chr8_+_66432475
Show fit
|
0.43 |
ENST00000415254.5
ENST00000396623.8
|
ADHFE1
|
alcohol dehydrogenase iron containing 1
|
|
chr15_-_77696142
Show fit
|
0.42 |
ENST00000561030.5
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chrX_+_153411472
Show fit
|
0.42 |
ENST00000338647.7
|
ZFP92
|
ZFP92 zinc finger protein
|
|
chr7_+_90709530
Show fit
|
0.41 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14
|
|
chr3_-_151249114
Show fit
|
0.41 |
ENST00000424796.6
|
P2RY14
|
purinergic receptor P2Y14
|
|
chr12_-_4649043
Show fit
|
0.40 |
ENST00000545990.6
ENST00000228850.6
|
AKAP3
|
A-kinase anchoring protein 3
|
|
chr3_+_46407558
Show fit
|
0.39 |
ENST00000357392.4
ENST00000400880.3
ENST00000433848.1
|
CCRL2
|
C-C motif chemokine receptor like 2
|
|
chr9_-_133609325
Show fit
|
0.39 |
ENST00000673969.1
|
FAM163B
|
family with sequence similarity 163 member B
|
|
chr1_+_81699665
Show fit
|
0.39 |
ENST00000359929.7
|
ADGRL2
|
adhesion G protein-coupled receptor L2
|
|
chr8_+_106726115
Show fit
|
0.38 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1
|
|
chr10_-_48251757
Show fit
|
0.38 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2
|
|
chr2_-_144430934
Show fit
|
0.38 |
ENST00000638087.1
ENST00000638007.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chrX_-_74614612
Show fit
|
0.37 |
ENST00000349225.2
ENST00000332687.11
|
RLIM
|
ring finger protein, LIM domain interacting
|
|
chr1_-_204166334
Show fit
|
0.37 |
ENST00000272190.9
|
REN
|
renin
|
|
chr17_+_31094880
Show fit
|
0.37 |
ENST00000487476.5
ENST00000356175.7
|
NF1
|
neurofibromin 1
|