Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for RFX3_RFX2
Z-value: 7.14
Transcription factors associated with RFX3_RFX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
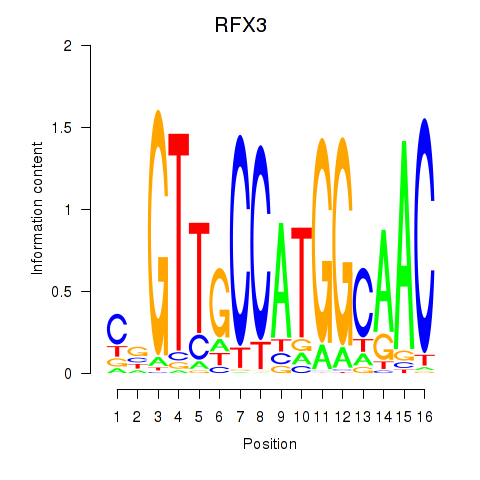
RFX3
|
ENSG00000080298.16 | regulatory factor X3 |
|
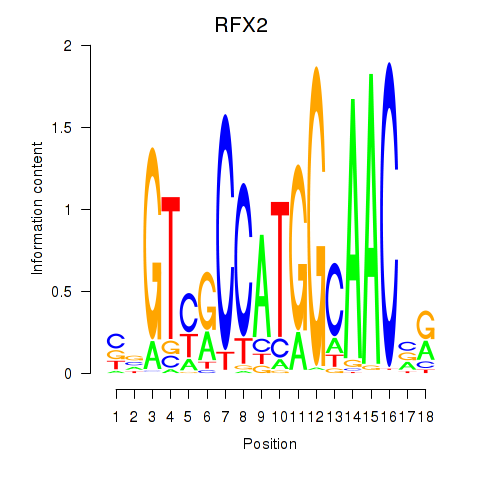
RFX2
|
ENSG00000087903.13 | regulatory factor X2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX2 | hg38_v1_chr19_-_6199510_6199564 | 0.65 | 5.5e-05 | Click! |
| RFX3 | hg38_v1_chr9_-_3489426_3489487 | 0.63 | 1.3e-04 | Click! |
Activity profile of RFX3_RFX2 motif
Sorted Z-values of RFX3_RFX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX3_RFX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 18.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 4.1 | 32.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 3.9 | 19.4 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 3.5 | 146.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 3.2 | 9.7 | GO:0015847 | putrescine transport(GO:0015847) |
| 3.0 | 9.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 3.0 | 8.9 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 2.9 | 11.5 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 2.7 | 13.4 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 2.4 | 7.3 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 2.1 | 8.4 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 2.0 | 10.0 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 2.0 | 29.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.8 | 27.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 1.8 | 7.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 1.8 | 7.1 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 1.8 | 3.5 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 1.7 | 15.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.6 | 22.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 1.6 | 14.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 1.6 | 11.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 1.6 | 9.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 1.6 | 29.6 | GO:0003341 | cilium movement(GO:0003341) |
| 1.5 | 65.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 1.5 | 7.6 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.5 | 31.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 1.5 | 14.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.4 | 20.7 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.4 | 15.0 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.3 | 5.3 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.3 | 24.9 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 1.2 | 3.6 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 1.2 | 3.6 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 1.2 | 14.0 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 1.1 | 9.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.1 | 7.9 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 1.1 | 7.6 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 1.1 | 8.6 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 1.1 | 9.6 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 1.0 | 3.1 | GO:0051041 | positive regulation of calcium-independent cell-cell adhesion(GO:0051041) |
| 1.0 | 10.4 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 1.0 | 6.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 1.0 | 20.1 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 1.0 | 11.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 1.0 | 4.9 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 1.0 | 5.8 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.9 | 16.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.9 | 6.0 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.8 | 5.7 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.8 | 13.0 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.8 | 0.8 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.8 | 4.8 | GO:0033504 | somite specification(GO:0001757) floor plate development(GO:0033504) |
| 0.8 | 11.1 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.8 | 6.2 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.8 | 6.9 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.7 | 5.2 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.7 | 8.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.7 | 17.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.6 | 1.9 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.6 | 5.5 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.6 | 26.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.6 | 7.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.6 | 1.7 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.5 | 2.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.5 | 4.3 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.5 | 1.6 | GO:0034092 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.5 | 5.9 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.5 | 5.5 | GO:0044782 | cilium organization(GO:0044782) |
| 0.5 | 1.5 | GO:0031393 | negative regulation of prostaglandin biosynthetic process(GO:0031393) |
| 0.5 | 27.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.5 | 26.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.5 | 19.5 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.5 | 1.0 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.5 | 2.8 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.5 | 3.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.5 | 11.4 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.5 | 3.8 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.5 | 1.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.5 | 18.1 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.4 | 9.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 | 3.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.4 | 4.3 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.4 | 2.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.4 | 3.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.4 | 7.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.4 | 1.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.4 | 1.1 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.4 | 7.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.4 | 19.9 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.4 | 1.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.3 | 1.0 | GO:0001928 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.3 | 7.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.3 | 3.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.3 | 4.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 2.0 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.3 | 1.6 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.3 | 4.0 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.3 | 32.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.3 | 3.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.3 | 17.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.3 | 3.6 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.3 | 1.2 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.3 | 5.8 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.3 | 3.7 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.3 | 1.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.3 | 0.8 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.3 | 1.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.3 | 2.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.3 | 4.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 13.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.3 | 0.8 | GO:0042245 | RNA repair(GO:0042245) |
| 0.3 | 11.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.2 | 0.7 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 6.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.2 | 31.9 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.2 | 1.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 4.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 5.6 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 16.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 34.5 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.2 | 1.8 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.2 | 7.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 0.4 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.2 | 4.3 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.2 | 4.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 6.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 4.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 0.8 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 4.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 15.8 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.2 | 2.8 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.2 | 13.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 0.9 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 1.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 33.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.2 | 4.7 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.2 | 1.0 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.2 | 12.4 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.2 | 4.9 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 | 39.8 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.2 | 2.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 23.0 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 9.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 16.7 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.2 | 8.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.2 | 4.6 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 8.3 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.2 | 8.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 19.7 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 6.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.4 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 6.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 9.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.1 | 3.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 3.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 10.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.9 | GO:0021522 | ventral spinal cord development(GO:0021517) spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 | 6.9 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 4.7 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 3.1 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.1 | 4.3 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) |
| 0.1 | 3.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 3.8 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 1.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 4.1 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 6.0 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.1 | 5.4 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 8.2 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 1.8 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.5 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.4 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 3.3 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 5.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.8 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.5 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 1.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.1 | 0.2 | GO:0006174 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 3.9 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 3.4 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 1.3 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 3.5 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 3.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 3.6 | GO:0017156 | calcium ion regulated exocytosis(GO:0017156) |
| 0.1 | 2.6 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 2.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 6.2 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 3.7 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.1 | 3.3 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.1 | 0.4 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.1 | 1.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.3 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.7 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 4.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 5.6 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 1.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 1.4 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 2.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 16.3 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 1.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 1.8 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 4.1 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 19.3 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 1.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 6.4 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 1.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.5 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 1.1 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.0 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 4.4 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 4.1 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 2.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 2.8 | GO:0006195 | purine nucleotide catabolic process(GO:0006195) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 3.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 2.8 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.5 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.8 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.6 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 2.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.3 | GO:0042308 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein import(GO:1904590) |
| 0.0 | 0.6 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 8.0 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 1.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.6 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.6 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 1.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.7 | GO:0030042 | actin filament depolymerization(GO:0030042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 26.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 7.8 | 62.2 | GO:0002177 | manchette(GO:0002177) |
| 6.6 | 26.2 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 6.2 | 18.7 | GO:0001534 | radial spoke(GO:0001534) |
| 3.7 | 11.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 3.4 | 37.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 3.0 | 9.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 2.0 | 6.0 | GO:0097545 | axonemal outer doublet(GO:0097545) |
| 1.9 | 9.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 1.4 | 48.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 1.4 | 14.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.3 | 25.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.2 | 7.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 1.1 | 131.7 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 1.0 | 6.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 1.0 | 5.7 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.9 | 9.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.9 | 6.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.8 | 5.7 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.8 | 15.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.8 | 39.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.8 | 5.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.7 | 13.5 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.7 | 4.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.7 | 20.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.7 | 16.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.6 | 3.8 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.6 | 112.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.6 | 11.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.6 | 6.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.5 | 6.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.5 | 14.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.5 | 8.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.5 | 4.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.5 | 11.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 4.8 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.4 | 6.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.4 | 5.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.4 | 7.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.4 | 9.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.4 | 6.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.4 | 2.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.4 | 29.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.4 | 3.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 1.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.3 | 3.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.3 | 31.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 4.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.3 | 2.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 123.2 | GO:0005929 | cilium(GO:0005929) |
| 0.3 | 19.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 7.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 22.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 0.7 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 9.8 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 2.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 36.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 126.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.2 | 16.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 6.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.2 | 4.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 1.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 3.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 7.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 0.6 | GO:0071920 | cleavage body(GO:0071920) |
| 0.2 | 1.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 19.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 6.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 12.0 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.9 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 22.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 7.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 13.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 12.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 17.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 7.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 5.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 3.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 3.5 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.6 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 15.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 11.1 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 3.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 3.8 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 67.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 0.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 24.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 8.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 4.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 6.5 | GO:1902911 | protein kinase complex(GO:1902911) |
| 0.0 | 1.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 15.7 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 2.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 11.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 3.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 5.4 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 3.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.0 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 4.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 2.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 13.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 19.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 4.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0097451 | astrocyte projection(GO:0097449) astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
| 0.0 | 2.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 4.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 3.2 | 9.7 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 2.5 | 25.5 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 2.5 | 10.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 2.3 | 7.0 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 2.3 | 9.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 2.3 | 29.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.9 | 5.6 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 1.8 | 31.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 1.8 | 5.3 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 1.6 | 6.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 1.5 | 22.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 1.4 | 5.8 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 1.2 | 13.4 | GO:0071253 | connexin binding(GO:0071253) |
| 1.2 | 3.6 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 1.1 | 8.6 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 1.1 | 5.3 | GO:0004802 | transketolase activity(GO:0004802) |
| 1.1 | 10.6 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 1.0 | 3.1 | GO:0031737 | CX3C chemokine receptor binding(GO:0031737) |
| 1.0 | 6.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 1.0 | 3.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.9 | 6.6 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.9 | 2.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.9 | 3.7 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 0.9 | 20.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.8 | 8.4 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.8 | 5.7 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.8 | 21.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.8 | 7.6 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.7 | 5.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.7 | 2.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.7 | 9.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.7 | 6.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.7 | 4.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.6 | 15.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.6 | 8.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.6 | 15.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.6 | 16.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.6 | 4.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.6 | 19.9 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.6 | 2.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.5 | 13.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.5 | 7.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.5 | 42.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.5 | 17.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.5 | 6.9 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.5 | 25.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 5.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 2.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.4 | 7.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.4 | 1.2 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.4 | 1.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.4 | 1.5 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.4 | 9.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.4 | 1.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.3 | 27.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 5.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 10.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 8.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 146.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.3 | 11.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 6.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 7.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 11.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 2.9 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.3 | 1.4 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 4.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 2.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.3 | 3.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.3 | 6.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 0.7 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 1.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 5.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 14.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 3.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 5.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 1.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 15.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.2 | 4.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 1.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 6.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.8 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 14.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 33.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 2.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 0.8 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.2 | 13.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 4.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 7.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 9.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 11.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 1.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 2.9 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 19.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 4.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 4.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 44.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 2.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 26.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 22.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 3.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 2.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.6 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 3.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 5.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 3.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 8.1 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 6.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 10.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 5.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 4.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 1.6 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.9 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 5.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 9.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 5.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 3.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 6.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.5 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 1.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 2.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 13.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 29.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.0 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 5.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 9.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 6.6 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 4.9 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 18.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 9.9 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 9.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.9 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 6.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 3.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 3.5 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 1.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.4 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 1.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 4.9 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 5.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 16.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 3.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.8 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 9.4 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 11.3 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 3.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 16.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 4.6 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 8.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 1.0 | GO:0002020 | protease binding(GO:0002020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 21.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.7 | 35.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.7 | 36.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.6 | 25.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.3 | 29.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.2 | 25.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 11.1 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 12.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 9.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 9.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 8.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 10.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 6.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 7.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 0.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 4.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 5.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 4.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 3.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 3.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 2.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 9.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 36.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.9 | 25.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.8 | 26.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.8 | 3.8 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.6 | 28.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 11.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.4 | 23.5 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.4 | 7.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 24.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 11.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 10.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 3.6 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.2 | 9.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 6.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 5.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 3.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 2.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 4.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 24.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 2.8 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.1 | 5.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 4.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 6.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 4.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 2.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 5.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 3.9 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 3.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 5.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 5.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.6 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 3.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 9.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 4.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 16.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 3.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 9.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.7 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 3.9 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 14.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |