Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for RFX7_RFX4_RFX1
Z-value: 5.38
Transcription factors associated with RFX7_RFX4_RFX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
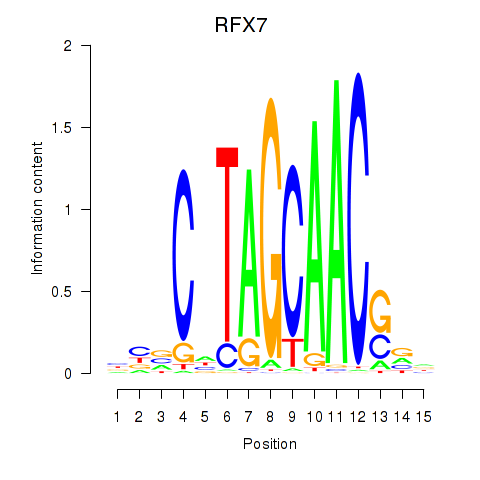
RFX7
|
ENSG00000181827.16 | regulatory factor X7 |
|
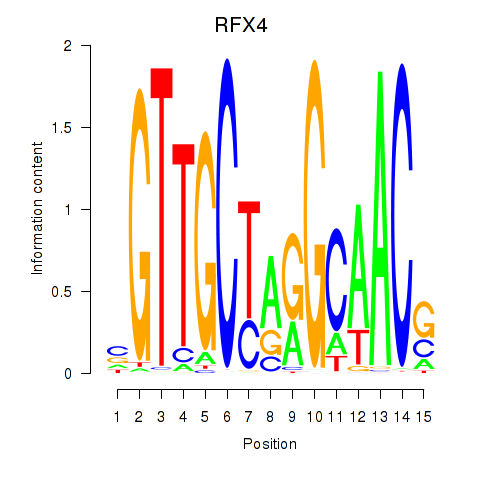
RFX4
|
ENSG00000111783.13 | regulatory factor X4 |
|
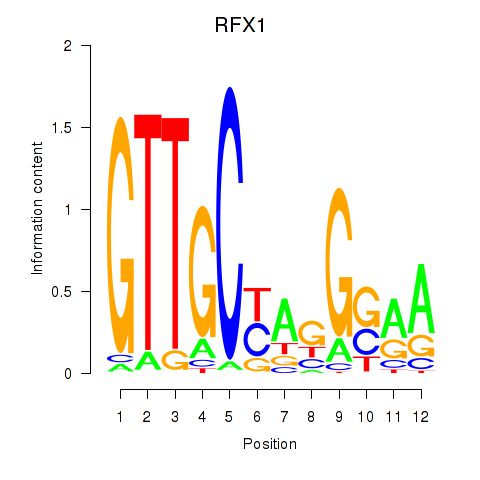
RFX1
|
ENSG00000132005.9 | regulatory factor X1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX4 | hg38_v1_chr12_+_106684696_106684755 | 0.72 | 3.4e-06 | Click! |
| RFX1 | hg38_v1_chr19_-_14006262_14006330 | 0.32 | 7.5e-02 | Click! |
| RFX7 | hg38_v1_chr15_-_56243829_56243891 | 0.19 | 3.0e-01 | Click! |
Activity profile of RFX7_RFX4_RFX1 motif
Sorted Z-values of RFX7_RFX4_RFX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX7_RFX4_RFX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 3.2 | 35.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 2.8 | 16.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 2.6 | 58.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 2.6 | 7.8 | GO:0051041 | positive regulation of calcium-independent cell-cell adhesion(GO:0051041) |
| 2.6 | 28.1 | GO:0003341 | cilium movement(GO:0003341) |
| 2.3 | 9.1 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 2.2 | 53.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 2.0 | 50.5 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) |
| 2.0 | 8.0 | GO:0090301 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 2.0 | 9.8 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 1.7 | 11.7 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 1.5 | 6.0 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 1.5 | 22.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 1.3 | 10.4 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 1.3 | 3.8 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 1.2 | 92.2 | GO:0042073 | intraciliary transport(GO:0042073) |
| 1.2 | 12.0 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 1.1 | 18.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 1.1 | 5.5 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 1.0 | 6.0 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 1.0 | 24.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.9 | 3.7 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.9 | 3.5 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.9 | 10.2 | GO:0044782 | cilium organization(GO:0044782) |
| 0.8 | 16.0 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.8 | 11.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.8 | 2.3 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.8 | 4.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.7 | 2.8 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.7 | 6.8 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.6 | 5.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.6 | 2.5 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.6 | 4.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.6 | 2.2 | GO:0009631 | cold acclimation(GO:0009631) |
| 0.5 | 5.3 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.5 | 6.0 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.5 | 3.5 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.5 | 1.4 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.4 | 0.8 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.4 | 1.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.4 | 3.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 1.8 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.4 | 1.8 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.4 | 1.8 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.3 | 3.7 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.3 | 3.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 5.7 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.3 | 6.2 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.3 | 5.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.3 | 0.9 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.3 | 1.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.3 | 2.5 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.3 | 14.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.3 | 1.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.3 | 1.6 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.3 | 4.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.2 | 4.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 0.7 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.2 | 1.0 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.2 | 1.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.2 | 2.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 | 1.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.2 | 1.7 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.2 | 2.7 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 32.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.2 | 0.6 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.2 | 1.5 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.2 | 0.8 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 0.4 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.2 | 0.9 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 35.2 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 1.6 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.2 | 1.2 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.2 | 11.3 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.2 | 2.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 1.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 2.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 1.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.2 | 2.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 5.2 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.2 | 2.8 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 0.8 | GO:0032213 | regulation of telomere maintenance via semi-conservative replication(GO:0032213) negative regulation of telomere maintenance via semi-conservative replication(GO:0032214) |
| 0.1 | 0.4 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 2.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 1.9 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.1 | 0.7 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.5 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.1 | 0.7 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 0.9 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 2.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.9 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 5.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.1 | 0.4 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.1 | 0.7 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 1.0 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.5 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.7 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 1.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.7 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 1.5 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 2.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 4.7 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 1.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 5.1 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 26.1 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.1 | 2.7 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.3 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 1.3 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.9 | GO:0036302 | atrioventricular canal development(GO:0036302) |
| 0.1 | 1.0 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 2.4 | GO:0016188 | synaptic vesicle maturation(GO:0016188) regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.7 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.9 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.2 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) |
| 0.1 | 1.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.4 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 1.1 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 1.3 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 3.0 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.6 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 4.9 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 0.6 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.6 | GO:0072092 | renal vesicle formation(GO:0072033) ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.1 | 2.8 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.1 | 0.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 1.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 1.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 1.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 34.5 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 0.5 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.6 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 4.2 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 0.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.3 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 1.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 2.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.6 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 1.7 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 3.8 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 1.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 2.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 1.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 8.6 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.7 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) lens induction in camera-type eye(GO:0060235) |
| 0.0 | 1.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 10.1 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 2.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 2.2 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.8 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.5 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 5.0 | GO:0006195 | purine nucleotide catabolic process(GO:0006195) |
| 0.0 | 0.6 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 2.1 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.5 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 1.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 1.6 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 1.1 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.5 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 4.0 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 1.5 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 1.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 1.2 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.8 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 3.6 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 7.0 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.3 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.8 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 3.2 | GO:0021915 | neural tube development(GO:0021915) |
| 0.0 | 1.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.2 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 18.1 | GO:0001534 | radial spoke(GO:0001534) |
| 5.2 | 57.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 5.2 | 41.4 | GO:0002177 | manchette(GO:0002177) |
| 3.9 | 23.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 3.2 | 9.6 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 2.0 | 10.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 1.7 | 5.2 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 1.7 | 5.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 1.5 | 19.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 1.4 | 8.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.3 | 14.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.3 | 62.1 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 1.3 | 3.9 | GO:0016939 | kinesin II complex(GO:0016939) |
| 1.1 | 16.5 | GO:0034464 | BBSome(GO:0034464) |
| 1.1 | 27.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 1.1 | 111.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 1.0 | 18.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.9 | 11.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.8 | 80.0 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.6 | 4.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.6 | 4.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.5 | 56.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.5 | 5.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 0.4 | GO:0071920 | cleavage body(GO:0071920) |
| 0.4 | 6.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 2.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 1.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 3.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 1.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 0.7 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 0.7 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.2 | 2.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 8.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 14.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 1.8 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 7.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 8.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 5.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 14.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.7 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 13.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 5.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 2.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 43.9 | GO:0005929 | cilium(GO:0005929) |
| 0.1 | 2.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 1.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 0.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 3.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 2.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 4.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 8.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 4.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.2 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 0.8 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 3.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 7.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 1.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 3.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 40.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 2.6 | 7.8 | GO:0031737 | CX3C chemokine receptor binding(GO:0031737) |
| 1.9 | 5.7 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.9 | 5.6 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 1.8 | 9.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.8 | 16.0 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 1.7 | 11.8 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 1.5 | 52.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 1.3 | 8.8 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 1.2 | 18.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.0 | 5.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.0 | 3.0 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.9 | 27.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.9 | 11.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.9 | 7.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.7 | 21.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.6 | 4.3 | GO:0097108 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.6 | 4.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.5 | 12.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.5 | 5.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 1.5 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.5 | 5.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 4.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.4 | 4.3 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.3 | 3.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.3 | 18.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 3.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 2.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.3 | 2.9 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.3 | 11.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 2.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 2.3 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.2 | 2.4 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 2.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 1.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 11.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 1.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 1.9 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.7 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 4.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 4.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 13.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 6.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 3.7 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.2 | 7.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 2.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 3.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 2.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 0.8 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 12.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 0.6 | GO:0030379 | neurotensin receptor activity, non-G-protein coupled(GO:0030379) |
| 0.2 | 2.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 2.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 7.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 6.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 1.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 12.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 16.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.5 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 1.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.4 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.7 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 8.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.3 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 2.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.8 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 1.4 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 1.0 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 1.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.7 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 2.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.2 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.1 | 0.2 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.1 | 5.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.4 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 9.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 2.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 2.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 22.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 21.1 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.6 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.1 | 2.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.7 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 4.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 1.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 2.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 10.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 7.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 1.9 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 3.0 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 4.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 2.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 3.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 3.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 4.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 3.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 3.9 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 5.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 15.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 4.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 22.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.5 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.4 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 7.7 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 14.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 5.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 14.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 4.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 3.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 5.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 5.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 6.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 3.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 3.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 4.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 8.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 6.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 4.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.8 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 3.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 1.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 3.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 7.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 3.2 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.1 | 2.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 9.5 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 2.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 3.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 3.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 5.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 4.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 4.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 1.5 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |