Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for RUNX3_BCL11A
Z-value: 3.30
Transcription factors associated with RUNX3_BCL11A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
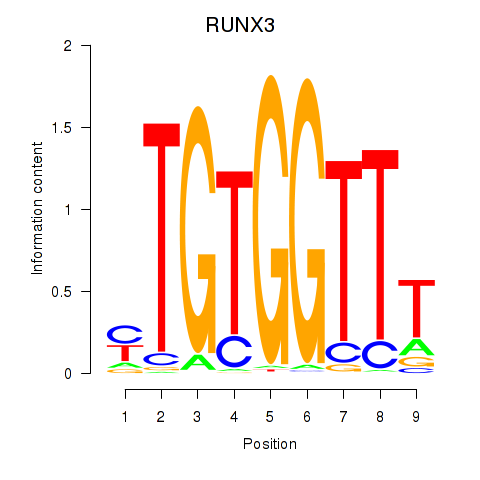
RUNX3
|
ENSG00000020633.19 | RUNX family transcription factor 3 |
|
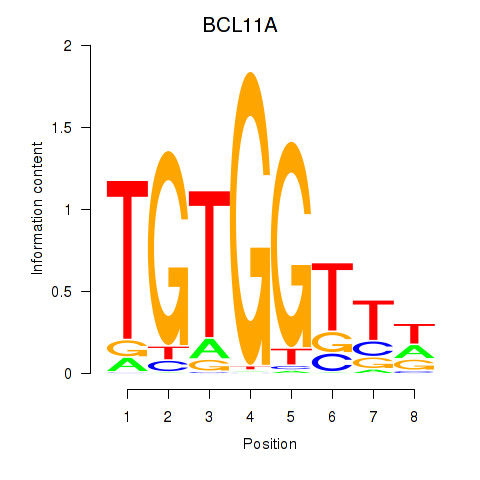
BCL11A
|
ENSG00000119866.22 | BAF chromatin remodeling complex subunit BCL11A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RUNX3 | hg38_v1_chr1_-_24930263_24930282 | 0.95 | 6.8e-17 | Click! |
| BCL11A | hg38_v1_chr2_-_60553409_60553461 | 0.55 | 1.1e-03 | Click! |
Activity profile of RUNX3_BCL11A motif
Sorted Z-values of RUNX3_BCL11A motif
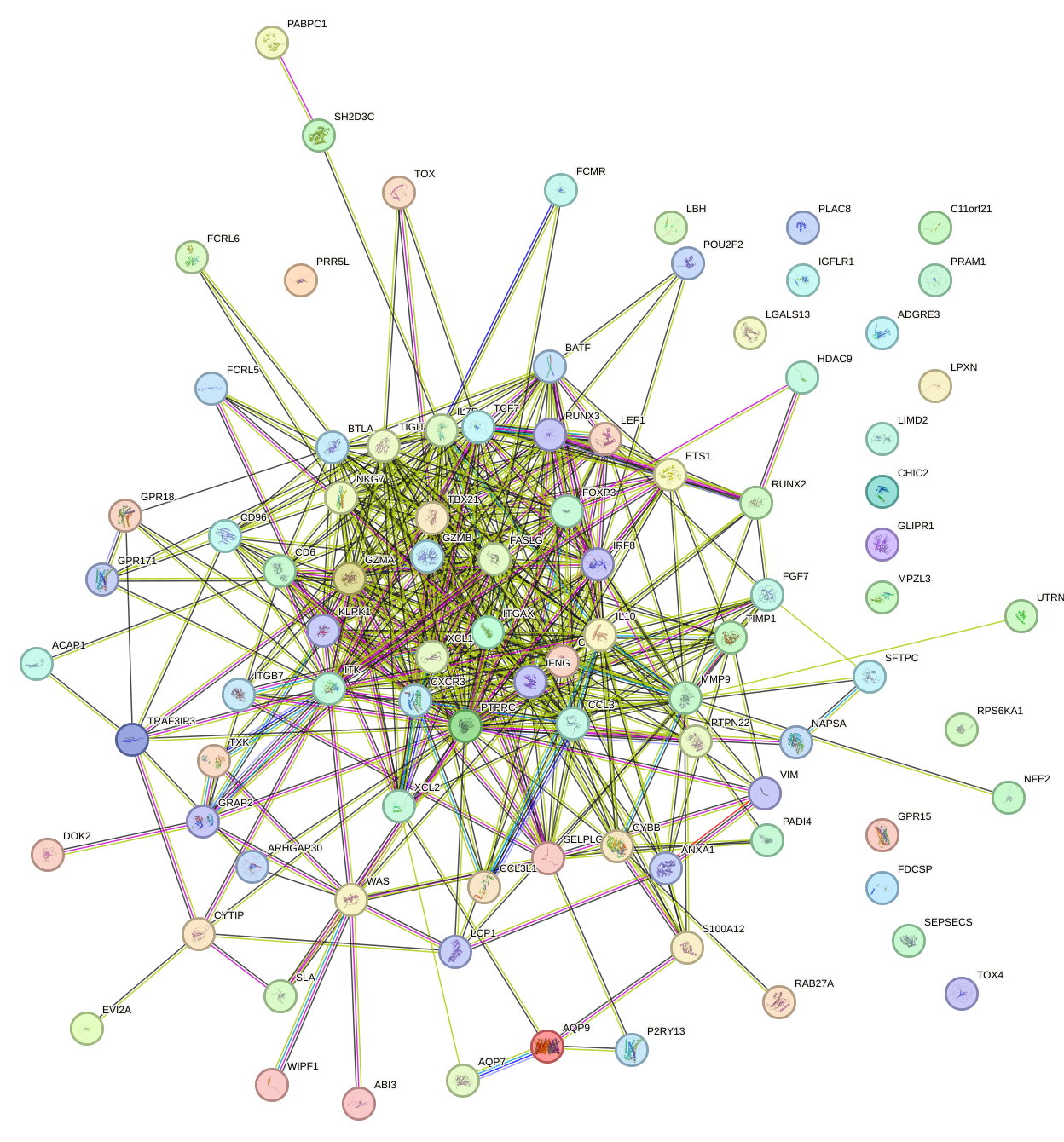
Network of associatons between targets according to the STRING database.
First level regulatory network of RUNX3_BCL11A
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.6 | 28.9 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 5.2 | 20.7 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 4.8 | 4.8 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 4.5 | 13.6 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 4.2 | 20.9 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 3.9 | 11.7 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 3.9 | 15.6 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 3.5 | 6.9 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 3.4 | 10.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 3.2 | 9.6 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 2.9 | 20.3 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 2.6 | 34.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 2.6 | 7.8 | GO:0036292 | DNA rewinding(GO:0036292) |
| 2.3 | 11.3 | GO:2000503 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 2.2 | 24.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 2.1 | 16.7 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) embryonic hindgut morphogenesis(GO:0048619) |
| 2.0 | 17.9 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 2.0 | 9.8 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 1.9 | 3.9 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 1.9 | 13.3 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.8 | 8.9 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 1.7 | 5.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 1.7 | 28.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 1.6 | 11.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 1.6 | 11.0 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 1.4 | 20.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.4 | 4.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 1.3 | 6.6 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 1.3 | 6.5 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 1.3 | 20.7 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 1.3 | 9.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 1.3 | 14.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 1.3 | 19.0 | GO:0032074 | regulation of endodeoxyribonuclease activity(GO:0032071) negative regulation of nuclease activity(GO:0032074) |
| 1.3 | 6.3 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 1.2 | 29.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 1.1 | 3.3 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 1.1 | 5.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 1.0 | 6.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 1.0 | 2.9 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 1.0 | 7.8 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.9 | 3.8 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.9 | 8.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.9 | 2.8 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.9 | 2.7 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.9 | 2.7 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.8 | 10.9 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.8 | 2.5 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 0.8 | 17.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.8 | 2.3 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.8 | 3.1 | GO:1901074 | cellular response to interleukin-13(GO:0035963) regulation of engulfment of apoptotic cell(GO:1901074) lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.8 | 16.6 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.7 | 4.4 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.7 | 2.9 | GO:0052417 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.7 | 3.5 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.7 | 2.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.7 | 2.7 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.7 | 19.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.7 | 2.6 | GO:2000734 | regulation of cell proliferation involved in mesonephros development(GO:2000606) negative regulation of cell proliferation involved in mesonephros development(GO:2000607) fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000699) glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000701) regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000702) negative regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000703) regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000733) negative regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000734) |
| 0.6 | 9.0 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.6 | 4.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.6 | 1.8 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.6 | 5.7 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.6 | 7.3 | GO:1902563 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.5 | 9.9 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.5 | 8.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.5 | 4.7 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.5 | 8.9 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.5 | 15.0 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.5 | 1.5 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.5 | 2.5 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.5 | 1.5 | GO:0070377 | negative regulation of ERK5 cascade(GO:0070377) |
| 0.5 | 2.4 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.5 | 1.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.5 | 2.8 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.5 | 24.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.5 | 10.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.4 | 1.8 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.4 | 3.9 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.4 | 1.7 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.4 | 1.3 | GO:0060690 | positive regulation of vascular wound healing(GO:0035470) epithelial cell differentiation involved in salivary gland development(GO:0060690) epithelial cell maturation involved in salivary gland development(GO:0060691) regulation of plasma cell differentiation(GO:1900098) positive regulation of plasma cell differentiation(GO:1900100) positive regulation of lactation(GO:1903489) |
| 0.4 | 1.3 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.4 | 1.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 20.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.4 | 0.8 | GO:1905237 | response to cyclosporin A(GO:1905237) |
| 0.4 | 10.4 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.4 | 5.9 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.4 | 1.8 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.4 | 2.8 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) common myeloid progenitor cell proliferation(GO:0035726) |
| 0.3 | 1.7 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.3 | 1.0 | GO:0021658 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.3 | 2.0 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.3 | 1.3 | GO:0032599 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.3 | 2.7 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 5.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 4.0 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 33.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.3 | 10.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 2.7 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 2.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 1.4 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.2 | 0.7 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.2 | 0.9 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.2 | 6.6 | GO:0072678 | T cell migration(GO:0072678) |
| 0.2 | 21.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 2.0 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.2 | 2.0 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.2 | 3.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.2 | 4.0 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 2.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 1.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.6 | GO:0060957 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.2 | 0.6 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 1.7 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.2 | 3.1 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.2 | 1.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 8.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.2 | 32.9 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.2 | 1.0 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 | 1.5 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.2 | 1.0 | GO:1905232 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) cellular response to L-glutamate(GO:1905232) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.2 | 1.1 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 4.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.2 | 2.4 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.1 | 2.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.9 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 1.0 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.1 | 1.8 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 1.4 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 3.7 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.1 | 5.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 3.1 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.1 | 3.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.3 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 1.5 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.1 | 7.7 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 2.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 2.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.5 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 1.6 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 1.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 7.8 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 2.4 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 0.4 | GO:1901072 | glucosamine-containing compound catabolic process(GO:1901072) |
| 0.1 | 0.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 2.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.6 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.8 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 1.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 4.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 8.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.7 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.6 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 0.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.3 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.1 | 1.3 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 1.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 5.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 0.8 | GO:1903800 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 1.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.5 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 1.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.9 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 1.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 1.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 4.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 2.0 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 1.8 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.5 | GO:0098909 | maintenance of centrosome location(GO:0051661) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.5 | GO:0035878 | ectodermal cell differentiation(GO:0010668) nail development(GO:0035878) |
| 0.0 | 7.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 38.5 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 3.3 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.5 | GO:1901256 | interleukin-21 production(GO:0032625) macrophage colony-stimulating factor production(GO:0036301) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.0 | 0.5 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.4 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 1.0 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 2.1 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 1.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 5.1 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.3 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.3 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 5.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 2.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.7 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 3.9 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.2 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.8 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 11.1 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.3 | GO:0050856 | regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.0 | 0.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.7 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.6 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 14.2 | GO:0071757 | hexameric IgM immunoglobulin complex(GO:0071757) |
| 1.8 | 8.9 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 1.7 | 20.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.5 | 21.5 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 1.1 | 7.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.1 | 21.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 1.0 | 34.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.8 | 4.2 | GO:0097059 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.8 | 15.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.7 | 2.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.7 | 2.9 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.7 | 27.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.7 | 13.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.7 | 44.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.4 | 9.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.4 | 14.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.3 | 36.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 6.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 7.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.3 | 67.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 9.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 6.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 3.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 10.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 4.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 23.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 3.0 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 3.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 63.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.3 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 0.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 1.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.5 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 1.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 12.7 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 25.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 8.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 0.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 4.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 9.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 0.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 2.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 37.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 3.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 5.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 12.3 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 7.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 6.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 10.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 6.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 5.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.2 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 2.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 8.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 1.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 4.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 2.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 4.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.7 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 1.0 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 14.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 4.4 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 4.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 2.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 27.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 3.5 | 31.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 2.8 | 11.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 2.0 | 14.2 | GO:0019862 | IgA binding(GO:0019862) |
| 2.0 | 9.8 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 1.3 | 10.7 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.3 | 6.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 1.3 | 20.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 1.3 | 6.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.2 | 9.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.2 | 10.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.2 | 29.9 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 1.0 | 3.1 | GO:0005139 | interleukin-7 receptor binding(GO:0005139) |
| 0.9 | 3.8 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.9 | 2.8 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.9 | 12.8 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.9 | 2.7 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.8 | 8.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.8 | 2.3 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.8 | 3.1 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.7 | 5.9 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.7 | 3.5 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.7 | 2.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.6 | 6.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.6 | 13.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.6 | 13.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.6 | 23.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.6 | 61.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.5 | 8.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.5 | 7.7 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.5 | 8.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.5 | 22.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 16.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.5 | 10.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.4 | 2.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 13.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.4 | 3.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.4 | 10.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 15.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.4 | 1.4 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.3 | 10.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.3 | 2.6 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.3 | 9.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 2.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.3 | 15.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 1.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 2.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 8.8 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.3 | 2.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 3.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 20.0 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 2.4 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 1.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 1.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 7.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 1.0 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.2 | 0.8 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 2.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 0.9 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 2.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 0.9 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 7.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 1.5 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.2 | 4.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 59.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 3.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 3.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 3.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 0.8 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 2.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 2.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 2.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 4.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 2.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.7 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.9 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 5.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 20.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.4 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.1 | 15.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 3.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.9 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.7 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 2.8 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.8 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.4 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 3.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 3.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 4.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 3.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 44.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 2.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 1.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 12.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.8 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 2.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.3 | GO:0070008 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 9.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 23.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 1.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 4.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.9 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 2.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 2.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 10.1 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 2.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 5.1 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 23.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.7 | 8.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.7 | 83.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.6 | 24.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.5 | 36.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.5 | 42.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.5 | 53.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.4 | 12.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.4 | 7.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.3 | 17.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.3 | 12.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.3 | 13.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.3 | 20.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 35.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.2 | 9.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 18.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 13.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 2.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 8.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 6.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 17.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 4.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 7.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 9.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 8.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 23.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 30.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 6.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 4.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 3.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 13.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 1.2 | 67.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.8 | 20.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.7 | 22.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.6 | 46.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.6 | 9.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 25.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.4 | 13.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 12.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 7.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.3 | 14.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 36.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 9.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 17.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 5.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 7.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 6.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 2.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 5.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 9.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 2.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 16.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 7.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 2.3 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 12.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 13.5 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 1.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 3.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 7.2 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 2.7 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 4.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 10.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 3.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 20.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 14.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 4.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 3.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 7.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 8.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 7.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.4 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |