Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for SIX1_SIX3_SIX2
Z-value: 1.00
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
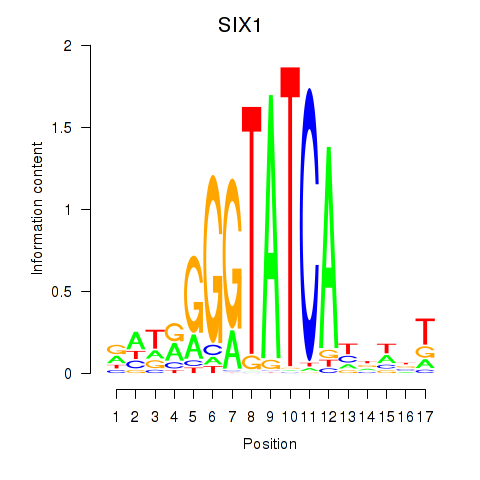
SIX1
|
ENSG00000126778.12 | SIX homeobox 1 |
|
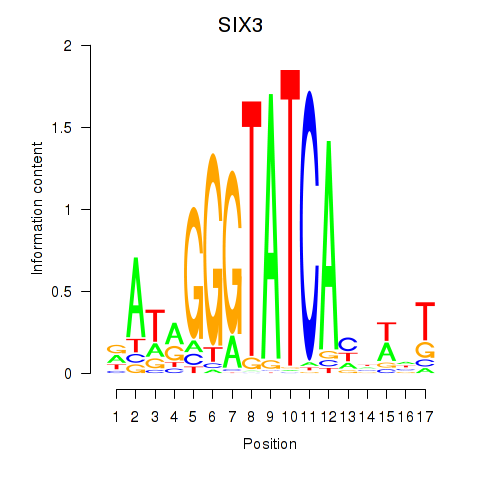
SIX3
|
ENSG00000138083.5 | SIX homeobox 3 |
|
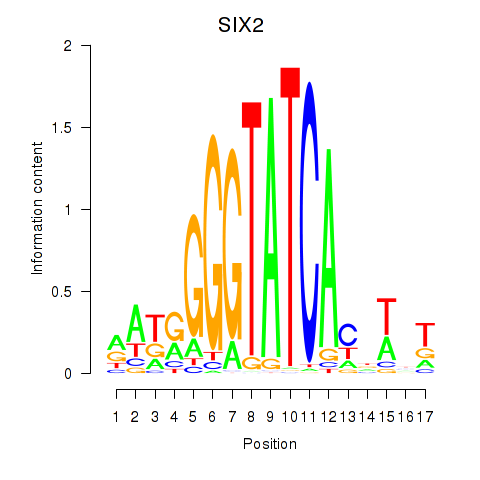
SIX2
|
ENSG00000170577.8 | SIX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX2 | hg38_v1_chr2_-_45009401_45009457 | -0.44 | 1.1e-02 | Click! |
| SIX1 | hg38_v1_chr14_-_60649449_60649490 | -0.42 | 1.6e-02 | Click! |
| SIX3 | hg38_v1_chr2_+_44941695_44941708 | 0.11 | 5.4e-01 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_58731936 | 8.62 |
ENST00000344743.8
|
GLYAT
|
glycine-N-acyltransferase |
| chr11_-_58731968 | 8.10 |
ENST00000278400.3
|
GLYAT
|
glycine-N-acyltransferase |
| chr10_+_5196831 | 6.25 |
ENST00000263126.3
|
AKR1C4
|
aldo-keto reductase family 1 member C4 |
| chr13_-_46105009 | 5.25 |
ENST00000439329.5
ENST00000674625.1 ENST00000181383.10 |
CPB2
|
carboxypeptidase B2 |
| chr3_-_42875871 | 3.36 |
ENST00000316161.6
ENST00000437102.1 |
CYP8B1
|
cytochrome P450 family 8 subfamily B member 1 |
| chr19_-_50333504 | 3.31 |
ENST00000474951.1
|
KCNC3
|
potassium voltage-gated channel subfamily C member 3 |
| chr2_+_233917371 | 3.04 |
ENST00000324695.9
ENST00000433712.6 |
TRPM8
|
transient receptor potential cation channel subfamily M member 8 |
| chr19_+_44942230 | 2.72 |
ENST00000592954.2
ENST00000589057.5 |
APOC4
APOC4-APOC2
|
apolipoprotein C4 APOC4-APOC2 readthrough (NMD candidate) |
| chr6_-_136526177 | 2.60 |
ENST00000617204.4
|
MAP7
|
microtubule associated protein 7 |
| chr4_+_154563003 | 2.47 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr6_-_136525961 | 2.36 |
ENST00000438100.6
|
MAP7
|
microtubule associated protein 7 |
| chr2_+_210556590 | 2.33 |
ENST00000233072.10
ENST00000619804.1 |
CPS1
|
carbamoyl-phosphate synthase 1 |
| chr6_-_136526472 | 2.32 |
ENST00000454590.5
ENST00000432797.6 |
MAP7
|
microtubule associated protein 7 |
| chrX_+_139530730 | 2.31 |
ENST00000218099.7
|
F9
|
coagulation factor IX |
| chr1_-_230745574 | 2.23 |
ENST00000681269.1
|
AGT
|
angiotensinogen |
| chr8_+_30638562 | 2.09 |
ENST00000517349.2
|
SMIM18
|
small integral membrane protein 18 |
| chr11_-_128842467 | 2.02 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr6_-_136526654 | 2.02 |
ENST00000611373.1
|
MAP7
|
microtubule associated protein 7 |
| chrX_+_139530752 | 1.92 |
ENST00000394090.2
|
F9
|
coagulation factor IX |
| chr1_+_241532370 | 1.89 |
ENST00000366559.9
ENST00000366557.8 |
KMO
|
kynurenine 3-monooxygenase |
| chr17_+_4771878 | 1.78 |
ENST00000270560.4
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr10_+_7703300 | 1.73 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_7703340 | 1.71 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr1_+_241532121 | 1.67 |
ENST00000366558.7
|
KMO
|
kynurenine 3-monooxygenase |
| chr8_-_29263063 | 1.59 |
ENST00000524189.6
|
KIF13B
|
kinesin family member 13B |
| chr7_-_99971845 | 1.58 |
ENST00000419575.1
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr3_+_149474688 | 1.55 |
ENST00000305354.5
ENST00000465758.1 |
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr1_+_209686173 | 1.45 |
ENST00000615289.4
ENST00000367028.6 ENST00000261465.5 |
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr2_+_233693659 | 1.44 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chr8_-_29263087 | 1.44 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr9_-_27005659 | 1.43 |
ENST00000380055.6
|
LRRC19
|
leucine rich repeat containing 19 |
| chr2_+_210477708 | 1.36 |
ENST00000673711.1
|
CPS1
|
carbamoyl-phosphate synthase 1 |
| chr14_-_94293024 | 1.32 |
ENST00000393096.5
|
SERPINA10
|
serpin family A member 10 |
| chr12_-_54984667 | 1.32 |
ENST00000524668.5
ENST00000533607.1 ENST00000449076.6 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr8_+_18391276 | 1.26 |
ENST00000286479.4
ENST00000520116.1 |
NAT2
|
N-acetyltransferase 2 |
| chr1_+_45500287 | 1.25 |
ENST00000401061.9
ENST00000616135.1 |
MMACHC
|
metabolism of cobalamin associated C |
| chr3_+_111999189 | 1.20 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr17_-_7177564 | 1.19 |
ENST00000570576.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr3_+_111998915 | 1.17 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr2_+_234050679 | 1.12 |
ENST00000373368.5
ENST00000168148.8 |
SPP2
|
secreted phosphoprotein 2 |
| chr3_+_111998739 | 1.11 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr3_+_111999326 | 1.09 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr1_+_196977550 | 1.07 |
ENST00000256785.5
|
CFHR5
|
complement factor H related 5 |
| chr1_+_207104226 | 1.01 |
ENST00000367070.8
|
C4BPA
|
complement component 4 binding protein alpha |
| chr11_-_124935973 | 1.01 |
ENST00000298251.5
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr11_-_34511710 | 1.00 |
ENST00000620316.4
ENST00000312319.6 |
ELF5
|
E74 like ETS transcription factor 5 |
| chr5_+_126423176 | 1.00 |
ENST00000542322.5
ENST00000544396.5 |
GRAMD2B
|
GRAM domain containing 2B |
| chr5_+_126423363 | 1.00 |
ENST00000285689.8
|
GRAMD2B
|
GRAM domain containing 2B |
| chr1_+_207104287 | 0.98 |
ENST00000421786.5
|
C4BPA
|
complement component 4 binding protein alpha |
| chr12_-_109021015 | 0.98 |
ENST00000546618.2
ENST00000610966.5 |
SVOP
|
SV2 related protein |
| chr10_-_101843920 | 0.98 |
ENST00000358038.7
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr11_-_49208589 | 0.97 |
ENST00000256999.7
|
FOLH1
|
folate hydrolase 1 |
| chr14_+_94581388 | 0.96 |
ENST00000554866.5
ENST00000556775.5 |
SERPINA5
|
serpin family A member 5 |
| chr14_-_94293260 | 0.95 |
ENST00000261994.9
|
SERPINA10
|
serpin family A member 10 |
| chr19_+_44942268 | 0.90 |
ENST00000591600.1
|
APOC4
|
apolipoprotein C4 |
| chr1_-_167935987 | 0.89 |
ENST00000367846.8
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr1_+_26410809 | 0.89 |
ENST00000254231.4
ENST00000326279.11 |
LIN28A
|
lin-28 homolog A |
| chr8_-_24956604 | 0.87 |
ENST00000610854.2
|
NEFL
|
neurofilament light |
| chr15_+_62561361 | 0.85 |
ENST00000561311.5
|
TLN2
|
talin 2 |
| chr5_+_126423122 | 0.85 |
ENST00000515200.5
|
GRAMD2B
|
GRAM domain containing 2B |
| chr1_+_50106265 | 0.84 |
ENST00000357083.8
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr17_-_46979240 | 0.84 |
ENST00000322329.5
|
RPRML
|
reprimo like |
| chr14_+_58244821 | 0.83 |
ENST00000216455.9
ENST00000412908.6 ENST00000557508.5 |
PSMA3
|
proteasome 20S subunit alpha 3 |
| chr5_-_35089620 | 0.80 |
ENST00000511486.5
ENST00000310101.9 ENST00000231423.7 ENST00000513753.5 ENST00000348262.7 |
PRLR
|
prolactin receptor |
| chr14_+_50560125 | 0.80 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr17_+_78187317 | 0.79 |
ENST00000409257.9
ENST00000591256.5 ENST00000589256.5 ENST00000588800.5 ENST00000591952.5 ENST00000327898.9 ENST00000586542.5 ENST00000586731.1 |
AFMID
|
arylformamidase |
| chr1_+_186296267 | 0.76 |
ENST00000533951.5
ENST00000367482.8 ENST00000635041.1 ENST00000367483.8 ENST00000367485.4 ENST00000445192.7 |
PRG4
|
proteoglycan 4 |
| chr3_+_179148099 | 0.75 |
ENST00000477735.1
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr2_+_234050732 | 0.75 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2 |
| chr3_+_42809439 | 0.74 |
ENST00000422265.6
ENST00000487368.4 |
ACKR2
ENSG00000273328.5
|
atypical chemokine receptor 2 novel transcript |
| chr11_+_64555956 | 0.74 |
ENST00000377581.7
|
SLC22A11
|
solute carrier family 22 member 11 |
| chr2_-_208190001 | 0.74 |
ENST00000451346.5
ENST00000341287.9 |
C2orf80
|
chromosome 2 open reading frame 80 |
| chr20_-_46684467 | 0.73 |
ENST00000372121.5
|
SLC13A3
|
solute carrier family 13 member 3 |
| chr6_-_43687774 | 0.73 |
ENST00000372133.8
ENST00000372116.5 ENST00000427312.1 |
MRPS18A
|
mitochondrial ribosomal protein S18A |
| chr5_+_140821598 | 0.72 |
ENST00000614258.1
ENST00000529859.2 ENST00000529619.5 |
PCDHA5
|
protocadherin alpha 5 |
| chr16_+_27203480 | 0.71 |
ENST00000286096.9
|
KDM8
|
lysine demethylase 8 |
| chr12_+_57230086 | 0.70 |
ENST00000414700.7
ENST00000557703.5 |
SHMT2
|
serine hydroxymethyltransferase 2 |
| chr2_+_127419686 | 0.69 |
ENST00000427769.5
|
PROC
|
protein C, inactivator of coagulation factors Va and VIIIa |
| chr1_+_161197372 | 0.69 |
ENST00000677550.1
ENST00000676600.1 ENST00000678507.1 ENST00000677579.1 ENST00000677231.1 ENST00000678911.1 ENST00000677846.1 |
NDUFS2
|
NADH:ubiquinone oxidoreductase core subunit S2 |
| chr4_+_2793077 | 0.67 |
ENST00000503393.8
|
SH3BP2
|
SH3 domain binding protein 2 |
| chr21_+_36069941 | 0.66 |
ENST00000530908.5
ENST00000439427.2 ENST00000290349.11 ENST00000399191.3 |
CBR1
|
carbonyl reductase 1 |
| chr2_-_60546162 | 0.66 |
ENST00000489183.1
ENST00000647469.1 ENST00000642180.1 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr10_+_5048748 | 0.66 |
ENST00000602997.5
ENST00000439082.7 |
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr3_+_188239858 | 0.65 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr20_+_38033719 | 0.65 |
ENST00000373433.9
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr22_+_30409097 | 0.65 |
ENST00000439838.5
ENST00000439023.3 |
ENSG00000249590.7
|
novel SEC14-like 2 (S. cerevisiae) (SEC14L2) and mitochondrial protein 18 kDa (MTP18) protein |
| chr4_+_88593251 | 0.63 |
ENST00000426683.5
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr10_+_4963406 | 0.63 |
ENST00000380872.9
ENST00000442997.5 |
AKR1C1
|
aldo-keto reductase family 1 member C1 |
| chr19_-_38229714 | 0.62 |
ENST00000416611.5
|
DPF1
|
double PHD fingers 1 |
| chr3_-_53846374 | 0.62 |
ENST00000315251.11
|
CHDH
|
choline dehydrogenase |
| chr8_-_133060347 | 0.62 |
ENST00000427060.6
|
SLA
|
Src like adaptor |
| chr8_+_75539893 | 0.61 |
ENST00000674002.1
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr11_+_61752603 | 0.60 |
ENST00000278836.10
|
MYRF
|
myelin regulatory factor |
| chr7_+_29479712 | 0.60 |
ENST00000412711.6
|
CHN2
|
chimerin 2 |
| chr7_+_151062547 | 0.60 |
ENST00000392826.6
ENST00000461735.1 |
SLC4A2
|
solute carrier family 4 member 2 |
| chr8_+_75539862 | 0.58 |
ENST00000396423.4
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr22_+_30425552 | 0.58 |
ENST00000629597.2
|
MTFP1
|
mitochondrial fission process 1 |
| chr2_-_222320124 | 0.58 |
ENST00000678139.1
|
ENSG00000288658.1
|
novel protein, ortholog of Gm2102 (M. musculus) |
| chr22_+_20967243 | 0.57 |
ENST00000683034.1
ENST00000440238.4 ENST00000405089.5 |
AIFM3
|
apoptosis inducing factor mitochondria associated 3 |
| chrX_+_153642473 | 0.55 |
ENST00000370167.8
|
DUSP9
|
dual specificity phosphatase 9 |
| chr6_-_35688907 | 0.55 |
ENST00000539068.5
ENST00000357266.9 |
FKBP5
|
FKBP prolyl isomerase 5 |
| chr9_+_12693327 | 0.54 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr1_+_176463164 | 0.54 |
ENST00000367661.7
ENST00000367662.5 |
PAPPA2
|
pappalysin 2 |
| chr12_-_117968223 | 0.54 |
ENST00000425217.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr7_-_72447061 | 0.53 |
ENST00000395276.6
ENST00000431984.5 |
CALN1
|
calneuron 1 |
| chr4_-_155376767 | 0.53 |
ENST00000450097.1
|
MAP9
|
microtubule associated protein 9 |
| chr19_+_18612848 | 0.52 |
ENST00000262817.8
|
TMEM59L
|
transmembrane protein 59 like |
| chr2_-_55269038 | 0.51 |
ENST00000417363.5
ENST00000412530.1 ENST00000366137.6 ENST00000420637.5 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr11_+_1409753 | 0.51 |
ENST00000524702.5
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr12_+_81078035 | 0.51 |
ENST00000261206.7
ENST00000548058.6 |
ACSS3
|
acyl-CoA synthetase short chain family member 3 |
| chr4_+_88593365 | 0.51 |
ENST00000452979.5
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr4_+_158210194 | 0.50 |
ENST00000508243.5
|
TMEM144
|
transmembrane protein 144 |
| chr19_-_14062028 | 0.50 |
ENST00000669674.2
|
PALM3
|
paralemmin 3 |
| chr10_-_101843765 | 0.49 |
ENST00000370046.5
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr15_+_90872862 | 0.48 |
ENST00000618099.4
|
FURIN
|
furin, paired basic amino acid cleaving enzyme |
| chr17_+_7627963 | 0.48 |
ENST00000575729.5
ENST00000340624.9 |
SHBG
|
sex hormone binding globulin |
| chr11_+_121576760 | 0.48 |
ENST00000532694.5
ENST00000534286.5 |
SORL1
|
sortilin related receptor 1 |
| chr19_-_11346486 | 0.47 |
ENST00000590482.5
|
TMEM205
|
transmembrane protein 205 |
| chr2_-_55269207 | 0.47 |
ENST00000263629.9
ENST00000403721.5 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr2_-_2324323 | 0.47 |
ENST00000648339.1
ENST00000647694.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr2_-_55269335 | 0.45 |
ENST00000441307.5
|
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr10_-_101843777 | 0.45 |
ENST00000356640.7
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr4_+_46993708 | 0.45 |
ENST00000513567.5
|
GABRB1
|
gamma-aminobutyric acid type A receptor subunit beta1 |
| chr15_+_58138368 | 0.45 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr2_-_73269483 | 0.45 |
ENST00000295133.9
|
FBXO41
|
F-box protein 41 |
| chr3_-_128052166 | 0.45 |
ENST00000648300.1
|
MGLL
|
monoglyceride lipase |
| chr16_-_4474549 | 0.44 |
ENST00000404295.7
ENST00000283429.11 ENST00000574425.5 |
NMRAL1
|
NmrA like redox sensor 1 |
| chr14_+_73097027 | 0.44 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr7_-_142933723 | 0.43 |
ENST00000442623.1
ENST00000265310.6 |
TRPV5
|
transient receptor potential cation channel subfamily V member 5 |
| chr1_-_13285154 | 0.43 |
ENST00000357367.6
ENST00000614831.1 |
PRAMEF8
|
PRAME family member 8 |
| chr12_+_57230301 | 0.43 |
ENST00000553474.5
|
SHMT2
|
serine hydroxymethyltransferase 2 |
| chr6_-_35688935 | 0.42 |
ENST00000542713.1
|
FKBP5
|
FKBP prolyl isomerase 5 |
| chr3_+_11226005 | 0.42 |
ENST00000413416.1
|
HRH1
|
histamine receptor H1 |
| chr4_-_46993520 | 0.42 |
ENST00000264318.4
|
GABRA4
|
gamma-aminobutyric acid type A receptor subunit alpha4 |
| chr8_-_18808837 | 0.42 |
ENST00000286485.12
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr3_+_35680994 | 0.42 |
ENST00000441454.5
|
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr3_+_35681352 | 0.41 |
ENST00000436702.5
ENST00000438071.1 |
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr12_+_20695469 | 0.41 |
ENST00000545604.5
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr2_+_148947868 | 0.41 |
ENST00000679129.1
|
KIF5C
|
kinesin family member 5C |
| chr6_+_150368997 | 0.40 |
ENST00000392255.7
ENST00000500320.7 ENST00000344419.8 |
IYD
|
iodotyrosine deiodinase |
| chr11_-_67401560 | 0.40 |
ENST00000679175.1
|
PPP1CA
|
protein phosphatase 1 catalytic subunit alpha |
| chr2_+_148978361 | 0.40 |
ENST00000678720.1
ENST00000678856.1 ENST00000677080.1 |
KIF5C
|
kinesin family member 5C |
| chr2_-_2324642 | 0.40 |
ENST00000650485.1
ENST00000649207.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr22_+_40678750 | 0.40 |
ENST00000249016.4
|
MCHR1
|
melanin concentrating hormone receptor 1 |
| chr5_-_161548124 | 0.39 |
ENST00000520240.5
ENST00000517901.5 ENST00000353437.10 |
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr16_-_66550142 | 0.39 |
ENST00000417693.8
ENST00000299697.12 ENST00000451102.7 |
TK2
|
thymidine kinase 2 |
| chr4_+_158210479 | 0.39 |
ENST00000504569.5
ENST00000509278.5 ENST00000514558.5 ENST00000503200.5 ENST00000296529.11 |
TMEM144
|
transmembrane protein 144 |
| chr3_+_25790076 | 0.39 |
ENST00000280701.8
ENST00000420173.2 |
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr4_-_69787955 | 0.39 |
ENST00000512870.1
|
SULT1B1
|
sulfotransferase family 1B member 1 |
| chr19_-_5867741 | 0.39 |
ENST00000252675.6
|
FUT5
|
fucosyltransferase 5 |
| chr6_+_167999092 | 0.39 |
ENST00000443060.6
|
KIF25
|
kinesin family member 25 |
| chr20_-_32657362 | 0.39 |
ENST00000360785.6
|
C20orf203
|
chromosome 20 open reading frame 203 |
| chr8_-_27605271 | 0.39 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr12_+_20695442 | 0.39 |
ENST00000540354.5
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr11_+_5422111 | 0.38 |
ENST00000300778.4
|
OR51Q1
|
olfactory receptor family 51 subfamily Q member 1 |
| chr9_+_74615582 | 0.38 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B |
| chr11_+_64555924 | 0.38 |
ENST00000301891.9
|
SLC22A11
|
solute carrier family 22 member 11 |
| chr12_+_20695553 | 0.37 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr6_-_138107412 | 0.37 |
ENST00000421351.4
|
PERP
|
p53 apoptosis effector related to PMP22 |
| chr4_-_174522791 | 0.37 |
ENST00000541923.5
ENST00000542498.5 |
HPGD
|
15-hydroxyprostaglandin dehydrogenase |
| chr10_-_112446932 | 0.37 |
ENST00000682195.1
ENST00000683505.1 ENST00000683895.1 ENST00000682839.1 ENST00000682055.1 |
ZDHHC6
|
zinc finger DHHC-type palmitoyltransferase 6 |
| chr4_-_155376876 | 0.37 |
ENST00000433024.5
ENST00000379248.6 |
MAP9
|
microtubule associated protein 9 |
| chr12_+_57230336 | 0.36 |
ENST00000555773.5
ENST00000554975.5 ENST00000449049.7 |
SHMT2
|
serine hydroxymethyltransferase 2 |
| chr4_+_2793056 | 0.36 |
ENST00000503219.5
|
SH3BP2
|
SH3 domain binding protein 2 |
| chr6_-_63319943 | 0.36 |
ENST00000622415.1
ENST00000370658.9 ENST00000485906.6 ENST00000370657.9 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr19_+_34926892 | 0.36 |
ENST00000303586.11
ENST00000601142.2 ENST00000439785.5 ENST00000601540.5 ENST00000601957.5 |
ZNF30
|
zinc finger protein 30 |
| chr22_+_20967212 | 0.36 |
ENST00000434714.6
|
AIFM3
|
apoptosis inducing factor mitochondria associated 3 |
| chr16_+_31108294 | 0.36 |
ENST00000287507.7
ENST00000394950.7 ENST00000219794.11 ENST00000561755.1 |
BCKDK
|
branched chain keto acid dehydrogenase kinase |
| chr6_+_72366730 | 0.36 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr3_+_6861107 | 0.36 |
ENST00000357716.9
ENST00000486284.5 ENST00000389336.8 |
GRM7
|
glutamate metabotropic receptor 7 |
| chr17_+_7654096 | 0.36 |
ENST00000577113.1
|
ATP1B2
|
ATPase Na+/K+ transporting subunit beta 2 |
| chr10_-_112446734 | 0.36 |
ENST00000684507.1
|
ZDHHC6
|
zinc finger DHHC-type palmitoyltransferase 6 |
| chr5_+_75337261 | 0.35 |
ENST00000680940.1
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr1_+_14929734 | 0.35 |
ENST00000376028.8
ENST00000400798.6 |
KAZN
|
kazrin, periplakin interacting protein |
| chr10_+_133394119 | 0.35 |
ENST00000317502.11
ENST00000432508.3 |
MTG1
|
mitochondrial ribosome associated GTPase 1 |
| chr1_+_13254212 | 0.35 |
ENST00000622421.2
|
PRAMEF5
|
PRAME family member 5 |
| chr4_-_155376935 | 0.35 |
ENST00000311277.9
|
MAP9
|
microtubule associated protein 9 |
| chr12_-_57742120 | 0.35 |
ENST00000257897.7
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr22_+_35717872 | 0.34 |
ENST00000249044.2
|
APOL5
|
apolipoprotein L5 |
| chr10_+_133394094 | 0.34 |
ENST00000477902.6
|
MTG1
|
mitochondrial ribosome associated GTPase 1 |
| chr15_+_89088417 | 0.34 |
ENST00000569550.5
ENST00000565066.5 ENST00000565973.5 ENST00000352732.10 |
ABHD2
|
abhydrolase domain containing 2, acylglycerol lipase |
| chr7_+_75882044 | 0.34 |
ENST00000428119.1
|
RHBDD2
|
rhomboid domain containing 2 |
| chr3_-_33097132 | 0.34 |
ENST00000307363.10
ENST00000307377.12 ENST00000440656.1 ENST00000436768.1 ENST00000342462.5 |
GLB1
TMPPE
|
galactosidase beta 1 transmembrane protein with metallophosphoesterase domain |
| chr2_+_208359686 | 0.34 |
ENST00000617735.4
|
PTH2R
|
parathyroid hormone 2 receptor |
| chr1_-_13201409 | 0.33 |
ENST00000625019.3
|
PRAMEF13
|
PRAME family member 13 |
| chr17_-_7627803 | 0.33 |
ENST00000573566.1
ENST00000269298.10 |
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr12_+_57230124 | 0.33 |
ENST00000553529.5
ENST00000554310.5 |
SHMT2
|
serine hydroxymethyltransferase 2 |
| chr10_-_5003850 | 0.33 |
ENST00000421196.7
ENST00000455190.2 ENST00000380753.8 |
AKR1C2
|
aldo-keto reductase family 1 member C2 |
| chr18_+_54828406 | 0.33 |
ENST00000262094.10
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr4_-_155376797 | 0.33 |
ENST00000650955.1
ENST00000515654.5 |
MAP9
|
microtubule associated protein 9 |
| chr20_+_18507520 | 0.33 |
ENST00000336714.8
ENST00000646240.1 ENST00000450074.6 ENST00000262544.6 |
SEC23B
|
SEC23 homolog B, COPII coat complex component |
| chr7_-_101200912 | 0.32 |
ENST00000440203.6
ENST00000379423.3 ENST00000223114.9 |
MOGAT3
|
monoacylglycerol O-acyltransferase 3 |
| chr2_+_32063588 | 0.31 |
ENST00000646571.1
|
SPAST
|
spastin |
| chr7_-_44159278 | 0.31 |
ENST00000345378.7
|
GCK
|
glucokinase |
| chr11_+_72189528 | 0.31 |
ENST00000312293.9
|
FOLR1
|
folate receptor alpha |
| chr11_+_925809 | 0.31 |
ENST00000332231.9
|
AP2A2
|
adaptor related protein complex 2 subunit alpha 2 |
| chr9_-_110256466 | 0.31 |
ENST00000374515.9
ENST00000374517.6 |
TXN
|
thioredoxin |
| chr1_+_197268222 | 0.31 |
ENST00000367400.8
ENST00000638467.1 ENST00000367399.6 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr6_+_101181254 | 0.30 |
ENST00000682090.1
ENST00000421544.6 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr21_+_43867215 | 0.30 |
ENST00000448287.5
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.3 | 7.9 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.2 | 3.7 | GO:1903717 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.7 | 3.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.6 | 18.8 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.4 | 4.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 3.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.4 | 3.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.4 | 2.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.4 | 1.4 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.3 | 0.9 | GO:1903937 | response to acrylamide(GO:1903937) |
| 0.3 | 1.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.2 | 3.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.2 | 2.0 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.2 | 1.3 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 0.7 | GO:0015744 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.2 | 0.5 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.2 | 2.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 0.7 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.2 | 1.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.2 | 0.5 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.2 | 0.6 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.9 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.8 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.7 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 1.6 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.9 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.5 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 1.4 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 1.0 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.7 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 1.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.3 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.1 | 0.4 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.1 | 0.4 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 1.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.8 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.3 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) |
| 0.1 | 0.3 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.1 | 0.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 1.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 1.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 1.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.6 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.2 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.3 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.4 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 1.7 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.4 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.1 | 3.4 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 8.3 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.1 | 0.3 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.1 | 0.3 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.4 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.1 | 0.4 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.3 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.7 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.7 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.4 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 1.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 1.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 2.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.3 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 2.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.7 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.4 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.7 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:0032445 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.4 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.4 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.4 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.6 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.4 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.5 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 0.3 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.5 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.0 | 0.9 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.9 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 1.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0046671 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 1.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.0 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.4 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 2.4 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 0.1 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.0 | 1.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 1.5 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 1.1 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 1.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.2 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0036027 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.2 | 2.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.9 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 3.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 2.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.3 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.1 | 6.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.3 | GO:1905103 | integral component of lysosomal membrane(GO:1905103) |
| 0.1 | 2.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.9 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 0.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.0 | 0.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 3.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 9.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 1.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 19.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.9 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 3.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 4.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 4.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 6.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 3.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 1.7 | 16.7 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.9 | 3.7 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.5 | 1.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.4 | 1.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.3 | 3.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 1.0 | GO:1904492 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.2 | 1.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 2.1 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 0.7 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.2 | 0.4 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 2.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 0.3 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.2 | 0.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 1.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 5.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.7 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 0.7 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.1 | 0.7 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 1.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.1 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.1 | 0.4 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 0.3 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 1.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.1 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 2.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.3 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.4 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.9 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.4 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.1 | 3.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.3 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.4 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 1.6 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 1.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.3 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.5 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 0.3 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.4 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.4 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.2 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.0 | 0.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 6.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.3 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 1.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 3.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.7 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.4 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 3.7 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 3.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 2.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 2.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 3.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.3 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 3.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 3.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 8.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 4.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 5.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 2.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 18.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 2.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 2.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 3.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 2.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 4.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |