Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for SMAD3
Z-value: 1.07
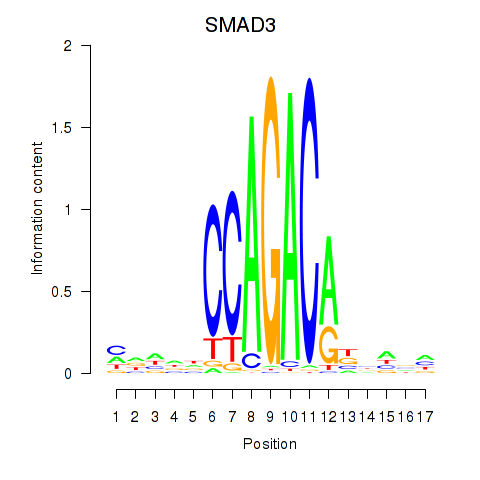
Transcription factors associated with SMAD3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD3
|
ENSG00000166949.17 | SMAD family member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD3 | hg38_v1_chr15_+_67067780_67067811 | -0.22 | 2.3e-01 | Click! |
Activity profile of SMAD3 motif
Sorted Z-values of SMAD3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_159714581 | 4.53 |
ENST00000255030.9
ENST00000437342.1 ENST00000368112.5 ENST00000368111.5 ENST00000368110.1 |
CRP
|
C-reactive protein |
| chr11_+_116829898 | 2.46 |
ENST00000227667.8
ENST00000375345.3 |
APOC3
|
apolipoprotein C3 |
| chr11_+_116830529 | 2.36 |
ENST00000630701.1
|
APOC3
|
apolipoprotein C3 |
| chr7_+_142656688 | 2.24 |
ENST00000390397.2
|
TRBV24-1
|
T cell receptor beta variable 24-1 |
| chr16_+_56669807 | 1.94 |
ENST00000332374.5
|
MT1H
|
metallothionein 1H |
| chr7_+_50308672 | 1.82 |
ENST00000439701.2
ENST00000438033.5 ENST00000492782.6 |
IKZF1
|
IKAROS family zinc finger 1 |
| chr5_+_132873660 | 1.80 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chr10_+_92691813 | 1.79 |
ENST00000472590.6
|
HHEX
|
hematopoietically expressed homeobox |
| chr1_-_160862880 | 1.70 |
ENST00000368034.9
|
CD244
|
CD244 molecule |
| chr1_-_160862700 | 1.67 |
ENST00000322302.7
ENST00000368033.7 |
CD244
|
CD244 molecule |
| chr7_+_142720652 | 1.63 |
ENST00000390400.2
|
TRBV28
|
T cell receptor beta variable 28 |
| chr16_-_3256587 | 1.63 |
ENST00000536379.5
ENST00000541159.5 ENST00000339854.8 ENST00000219596.6 |
MEFV
|
MEFV innate immuity regulator, pyrin |
| chr6_-_27132750 | 1.58 |
ENST00000607124.1
ENST00000339812.3 |
H2BC11
|
H2B clustered histone 11 |
| chr16_-_31065011 | 1.56 |
ENST00000539836.3
ENST00000535577.5 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr7_+_142469521 | 1.43 |
ENST00000390371.3
|
TRBV6-6
|
T cell receptor beta variable 6-6 |
| chr1_+_89821921 | 1.37 |
ENST00000394593.7
|
LRRC8D
|
leucine rich repeat containing 8 VRAC subunit D |
| chr16_+_30472700 | 1.36 |
ENST00000358164.9
|
ITGAL
|
integrin subunit alpha L |
| chr11_+_61102465 | 1.31 |
ENST00000347785.8
ENST00000544014.1 |
CD5
|
CD5 molecule |
| chr16_-_31064952 | 1.31 |
ENST00000426488.6
|
ZNF668
|
zinc finger protein 668 |
| chr19_-_40850442 | 1.30 |
ENST00000301141.10
|
CYP2A6
|
cytochrome P450 family 2 subfamily A member 6 |
| chr7_+_142492121 | 1.24 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6 |
| chr6_+_27133032 | 1.21 |
ENST00000359193.3
|
H2AC11
|
H2A clustered histone 11 |
| chr5_-_128339191 | 1.20 |
ENST00000507835.5
|
FBN2
|
fibrillin 2 |
| chr14_+_21868822 | 1.17 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr10_+_92691897 | 1.17 |
ENST00000492654.3
|
HHEX
|
hematopoietically expressed homeobox |
| chr19_+_18173148 | 1.17 |
ENST00000597802.2
|
IFI30
|
IFI30 lysosomal thiol reductase |
| chr6_-_27146841 | 1.17 |
ENST00000356950.2
|
H2BC12
|
H2B clustered histone 12 |
| chr7_+_139829242 | 1.16 |
ENST00000455353.6
ENST00000458722.6 ENST00000448866.7 ENST00000411653.6 |
TBXAS1
|
thromboxane A synthase 1 |
| chr16_+_30472733 | 1.12 |
ENST00000356798.11
ENST00000433423.2 |
ITGAL
|
integrin subunit alpha L |
| chr6_-_26123910 | 1.06 |
ENST00000314332.5
ENST00000396984.1 |
H2BC4
|
H2B clustered histone 4 |
| chr3_-_120647018 | 1.04 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr6_+_26124161 | 1.04 |
ENST00000377791.4
ENST00000602637.1 |
H2AC6
|
H2A clustered histone 6 |
| chr6_+_25726767 | 1.01 |
ENST00000274764.5
|
H2BC1
|
H2B clustered histone 1 |
| chr9_+_133534807 | 0.97 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS like 2 |
| chr7_+_142580911 | 0.91 |
ENST00000621184.1
|
TRBV12-5
|
T cell receptor beta variable 12-5 |
| chr7_+_100720463 | 0.89 |
ENST00000252723.3
|
EPO
|
erythropoietin |
| chr14_+_24130659 | 0.87 |
ENST00000267426.6
|
FITM1
|
fat storage inducing transmembrane protein 1 |
| chr19_+_51225059 | 0.86 |
ENST00000436584.6
ENST00000421133.6 ENST00000262262.5 ENST00000391796.7 |
CD33
|
CD33 molecule |
| chr15_+_81299416 | 0.85 |
ENST00000558332.3
|
IL16
|
interleukin 16 |
| chr1_-_225941383 | 0.84 |
ENST00000420304.6
|
LEFTY2
|
left-right determination factor 2 |
| chr1_-_225941212 | 0.83 |
ENST00000366820.10
|
LEFTY2
|
left-right determination factor 2 |
| chr14_+_92923143 | 0.82 |
ENST00000216492.10
ENST00000334654.4 |
CHGA
|
chromogranin A |
| chr6_+_26216928 | 0.82 |
ENST00000303910.4
|
H2AC8
|
H2A clustered histone 8 |
| chr18_+_13218195 | 0.80 |
ENST00000679167.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr7_+_142587857 | 0.79 |
ENST00000617639.1
|
TRBV14
|
T cell receptor beta variable 14 |
| chr19_+_41088450 | 0.78 |
ENST00000330436.4
|
CYP2A13
|
cytochrome P450 family 2 subfamily A member 13 |
| chr6_-_24809798 | 0.78 |
ENST00000562221.1
|
ENSG00000282804.1
|
novel protein |
| chr2_-_224982420 | 0.77 |
ENST00000645028.1
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr17_-_40937445 | 0.75 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr1_+_201190786 | 0.72 |
ENST00000335211.9
ENST00000295591.12 |
IGFN1
|
immunoglobulin like and fibronectin type III domain containing 1 |
| chr12_-_9760893 | 0.72 |
ENST00000228434.7
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr20_+_57351510 | 0.72 |
ENST00000411894.5
ENST00000429339.5 |
RAE1
|
ribonucleic acid export 1 |
| chr17_-_35868885 | 0.71 |
ENST00000604834.6
|
HEATR9
|
HEAT repeat containing 9 |
| chr6_+_139028680 | 0.71 |
ENST00000367660.4
|
ABRACL
|
ABRA C-terminal like |
| chr10_-_133358006 | 0.70 |
ENST00000278025.9
ENST00000368552.7 |
FUOM
|
fucose mutarotase |
| chr12_+_3491189 | 0.70 |
ENST00000382622.4
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr12_+_65169546 | 0.69 |
ENST00000308330.3
|
LEMD3
|
LEM domain containing 3 |
| chr11_-_65382632 | 0.68 |
ENST00000294187.10
ENST00000398802.6 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25 member 45 |
| chr12_+_67648737 | 0.68 |
ENST00000344096.4
ENST00000393555.3 |
DYRK2
|
dual specificity tyrosine phosphorylation regulated kinase 2 |
| chr9_+_133534697 | 0.67 |
ENST00000651351.2
|
ADAMTSL2
|
ADAMTS like 2 |
| chr16_+_2471284 | 0.67 |
ENST00000293973.2
|
NTN3
|
netrin 3 |
| chr20_-_64079479 | 0.66 |
ENST00000395042.2
|
RGS19
|
regulator of G protein signaling 19 |
| chr6_-_25726553 | 0.66 |
ENST00000297012.5
|
H2AC1
|
H2A clustered histone 1 |
| chr17_-_35868858 | 0.66 |
ENST00000603870.5
ENST00000603218.1 |
HEATR9
|
HEAT repeat containing 9 |
| chr20_+_57351218 | 0.65 |
ENST00000371242.6
ENST00000527947.5 ENST00000395841.7 |
RAE1
|
ribonucleic acid export 1 |
| chr8_+_22392821 | 0.64 |
ENST00000520832.1
|
SLC39A14
|
solute carrier family 39 member 14 |
| chrX_+_50067576 | 0.64 |
ENST00000376108.7
|
CLCN5
|
chloride voltage-gated channel 5 |
| chr4_-_176195563 | 0.63 |
ENST00000280191.7
|
SPATA4
|
spermatogenesis associated 4 |
| chr1_-_11858935 | 0.61 |
ENST00000376468.4
|
NPPB
|
natriuretic peptide B |
| chr2_-_128027273 | 0.58 |
ENST00000259235.7
ENST00000357702.9 ENST00000424298.5 |
SAP130
|
Sin3A associated protein 130 |
| chr1_-_160647287 | 0.58 |
ENST00000235739.6
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr17_+_4710622 | 0.57 |
ENST00000574954.5
ENST00000269260.7 ENST00000346341.6 ENST00000572457.5 ENST00000381488.10 ENST00000412477.7 ENST00000571428.5 ENST00000575877.5 |
ARRB2
|
arrestin beta 2 |
| chr7_+_142384328 | 0.55 |
ENST00000390361.3
|
TRBV7-3
|
T cell receptor beta variable 7-3 |
| chr2_+_219245455 | 0.55 |
ENST00000409638.7
ENST00000396738.7 ENST00000409516.7 |
STK16
|
serine/threonine kinase 16 |
| chr15_-_82647960 | 0.54 |
ENST00000615198.4
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr20_+_57351528 | 0.53 |
ENST00000395840.6
|
RAE1
|
ribonucleic acid export 1 |
| chr7_+_31687208 | 0.53 |
ENST00000409146.3
ENST00000342032.8 |
PPP1R17
|
protein phosphatase 1 regulatory subunit 17 |
| chr15_+_82262781 | 0.53 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000682753.1 ENST00000566861.5 ENST00000565432.1 |
SAXO2
|
stabilizer of axonemal microtubules 2 |
| chr6_+_47698538 | 0.52 |
ENST00000327753.7
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr2_-_49154507 | 0.51 |
ENST00000406846.7
|
FSHR
|
follicle stimulating hormone receptor |
| chr14_-_64942783 | 0.51 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2 |
| chr15_-_82647123 | 0.51 |
ENST00000569257.5
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr14_-_24081986 | 0.51 |
ENST00000560550.1
|
NRL
|
neural retina leucine zipper |
| chr8_+_10672623 | 0.51 |
ENST00000304519.10
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr17_-_41309253 | 0.50 |
ENST00000391352.1
|
KRTAP16-1
|
keratin associated protein 16-1 |
| chr6_-_27831557 | 0.50 |
ENST00000611927.2
|
H4C12
|
H4 clustered histone 12 |
| chr4_-_185775376 | 0.50 |
ENST00000456596.5
ENST00000414724.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_47698574 | 0.49 |
ENST00000283303.3
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr1_-_9069572 | 0.48 |
ENST00000377414.7
|
SLC2A5
|
solute carrier family 2 member 5 |
| chr15_-_82647503 | 0.48 |
ENST00000567678.1
ENST00000620182.4 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr1_-_9069797 | 0.48 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 member 5 |
| chr15_+_65530418 | 0.47 |
ENST00000562901.5
ENST00000261875.10 ENST00000442729.6 ENST00000565299.5 ENST00000568793.5 |
HACD3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr14_-_24081928 | 0.47 |
ENST00000396995.1
|
NRL
|
neural retina leucine zipper |
| chr5_+_140450 | 0.47 |
ENST00000502646.1
|
PLEKHG4B
|
pleckstrin homology and RhoGEF domain containing G4B |
| chr2_-_49154433 | 0.47 |
ENST00000454032.5
ENST00000304421.8 |
FSHR
|
follicle stimulating hormone receptor |
| chrX_-_48470243 | 0.47 |
ENST00000429543.2
ENST00000620913.5 |
SLC38A5
|
solute carrier family 38 member 5 |
| chrX_-_48469557 | 0.46 |
ENST00000488083.5
|
SLC38A5
|
solute carrier family 38 member 5 |
| chr1_-_88891496 | 0.46 |
ENST00000448623.5
ENST00000370500.10 ENST00000418217.1 |
GTF2B
|
general transcription factor IIB |
| chr18_+_46173495 | 0.46 |
ENST00000587591.5
ENST00000588730.1 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chrX_+_9560465 | 0.45 |
ENST00000647060.1
|
TBL1X
|
transducin beta like 1 X-linked |
| chr16_-_375205 | 0.44 |
ENST00000448854.1
|
PGAP6
|
post-glycosylphosphatidylinositol attachment to proteins 6 |
| chr11_-_32794610 | 0.44 |
ENST00000531481.1
ENST00000335185.10 |
CCDC73
|
coiled-coil domain containing 73 |
| chr22_+_41560973 | 0.44 |
ENST00000306149.12
|
CSDC2
|
cold shock domain containing C2 |
| chr20_+_57351569 | 0.44 |
ENST00000452119.1
|
RAE1
|
ribonucleic acid export 1 |
| chr6_-_26027274 | 0.43 |
ENST00000377745.4
|
H4C2
|
H4 clustered histone 2 |
| chr13_+_45464901 | 0.43 |
ENST00000349995.10
|
COG3
|
component of oligomeric golgi complex 3 |
| chr15_+_63504583 | 0.43 |
ENST00000380324.8
ENST00000561442.5 ENST00000560070.5 |
USP3
|
ubiquitin specific peptidase 3 |
| chr6_+_292050 | 0.42 |
ENST00000344450.9
|
DUSP22
|
dual specificity phosphatase 22 |
| chr3_+_51707059 | 0.42 |
ENST00000395052.8
|
GRM2
|
glutamate metabotropic receptor 2 |
| chr19_-_55180104 | 0.42 |
ENST00000537500.5
|
SYT5
|
synaptotagmin 5 |
| chr17_-_40772162 | 0.42 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr11_+_5691004 | 0.41 |
ENST00000414641.5
|
TRIM22
|
tripartite motif containing 22 |
| chr11_-_64744993 | 0.41 |
ENST00000377485.5
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr7_+_142399860 | 0.40 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1 |
| chr1_+_59297057 | 0.40 |
ENST00000303721.12
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr6_+_27824084 | 0.40 |
ENST00000355057.3
|
H4C11
|
H4 clustered histone 11 |
| chr5_-_133026533 | 0.40 |
ENST00000509437.6
ENST00000355372.6 ENST00000513541.5 ENST00000509008.1 ENST00000513848.5 ENST00000504170.2 ENST00000324170.7 |
ZCCHC10
|
zinc finger CCHC-type containing 10 |
| chr22_-_35617321 | 0.40 |
ENST00000397326.7
ENST00000442617.1 |
MB
|
myoglobin |
| chr4_+_69051362 | 0.39 |
ENST00000502942.5
|
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr1_+_59296971 | 0.39 |
ENST00000371218.8
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr2_-_24793382 | 0.38 |
ENST00000328379.6
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr14_-_64942720 | 0.38 |
ENST00000557049.1
ENST00000389614.6 |
GPX2
|
glutathione peroxidase 2 |
| chr3_-_52897541 | 0.38 |
ENST00000355083.11
ENST00000504329.1 |
STIMATE
STIMATE-MUSTN1
|
STIM activating enhancer STIMATE-MUSTN1 readthrough |
| chr11_-_72434330 | 0.37 |
ENST00000542555.2
ENST00000535990.5 ENST00000294053.9 ENST00000538039.6 ENST00000340729.9 |
CLPB
|
caseinolytic mitochondrial matrix peptidase chaperone subunit B |
| chr12_+_67269328 | 0.37 |
ENST00000545606.6
|
CAND1
|
cullin associated and neddylation dissociated 1 |
| chr13_+_24251686 | 0.37 |
ENST00000434675.5
ENST00000494772.5 |
SPATA13
|
spermatogenesis associated 13 |
| chr18_+_13611764 | 0.36 |
ENST00000585931.5
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr2_-_128028114 | 0.36 |
ENST00000259234.10
|
SAP130
|
Sin3A associated protein 130 |
| chr11_-_72070050 | 0.36 |
ENST00000535087.5
ENST00000535838.5 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr15_+_63504511 | 0.36 |
ENST00000540797.5
|
USP3
|
ubiquitin specific peptidase 3 |
| chrX_-_27463341 | 0.35 |
ENST00000412172.4
|
PPP4R3C
|
protein phosphatase 4 regulatory subunit 3C |
| chr2_-_128028010 | 0.35 |
ENST00000643581.2
ENST00000450957.1 |
SAP130
|
Sin3A associated protein 130 |
| chr3_+_125969214 | 0.34 |
ENST00000508088.1
|
ROPN1B
|
rhophilin associated tail protein 1B |
| chr9_-_34637719 | 0.34 |
ENST00000378892.5
ENST00000680277.1 ENST00000277010.9 ENST00000679597.1 ENST00000680244.1 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr9_-_34637800 | 0.34 |
ENST00000680730.1
ENST00000477726.1 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr15_-_28712323 | 0.34 |
ENST00000563027.1
|
GOLGA8M
|
golgin A8 family member M |
| chr5_+_55851349 | 0.34 |
ENST00000652347.2
|
IL31RA
|
interleukin 31 receptor A |
| chr6_+_26240385 | 0.33 |
ENST00000244537.6
|
H4C6
|
H4 clustered histone 6 |
| chr6_+_30883085 | 0.33 |
ENST00000504651.5
ENST00000512694.5 ENST00000515233.5 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr6_+_146598961 | 0.33 |
ENST00000522242.5
ENST00000397944.8 |
ADGB
|
androglobin |
| chr6_-_52577012 | 0.33 |
ENST00000182527.4
|
TRAM2
|
translocation associated membrane protein 2 |
| chr19_-_42220109 | 0.32 |
ENST00000595337.5
|
DEDD2
|
death effector domain containing 2 |
| chr12_+_49741802 | 0.32 |
ENST00000423828.5
ENST00000550445.5 |
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr11_-_83724922 | 0.32 |
ENST00000434967.1
ENST00000530800.5 |
DLG2
|
discs large MAGUK scaffold protein 2 |
| chr22_-_41281621 | 0.31 |
ENST00000452543.5
ENST00000418067.1 |
RANGAP1
|
Ran GTPase activating protein 1 |
| chr15_+_67825504 | 0.31 |
ENST00000380035.3
ENST00000554054.5 |
SKOR1
|
SKI family transcriptional corepressor 1 |
| chr1_+_153217578 | 0.31 |
ENST00000368744.4
|
PRR9
|
proline rich 9 |
| chr19_-_55180010 | 0.30 |
ENST00000589172.5
|
SYT5
|
synaptotagmin 5 |
| chr17_-_1078959 | 0.29 |
ENST00000571945.5
ENST00000536794.6 |
ABR
|
ABR activator of RhoGEF and GTPase |
| chr1_-_60073750 | 0.29 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr1_+_112718880 | 0.28 |
ENST00000361886.4
|
TAFA3
|
TAFA chemokine like family member 3 |
| chr11_-_5441514 | 0.28 |
ENST00000380211.1
|
OR51I1
|
olfactory receptor family 51 subfamily I member 1 |
| chr6_+_144659814 | 0.28 |
ENST00000367525.3
|
UTRN
|
utrophin |
| chrX_+_49829260 | 0.28 |
ENST00000376141.5
ENST00000218068.7 |
PAGE4
|
PAGE family member 4 |
| chr3_-_14178615 | 0.27 |
ENST00000511155.1
|
XPC
|
XPC complex subunit, DNA damage recognition and repair factor |
| chr6_+_116616467 | 0.27 |
ENST00000229554.10
ENST00000368581.8 ENST00000368580.4 |
RSPH4A
|
radial spoke head component 4A |
| chr11_-_133532493 | 0.26 |
ENST00000524381.6
|
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr11_+_124954108 | 0.26 |
ENST00000529051.5
|
CCDC15
|
coiled-coil domain containing 15 |
| chr21_+_31659666 | 0.26 |
ENST00000389995.4
ENST00000270142.11 |
SOD1
|
superoxide dismutase 1 |
| chr18_+_32018817 | 0.26 |
ENST00000217740.4
ENST00000583184.1 |
RNF125
ENSG00000263917.1
|
ring finger protein 125 novel transcript |
| chr12_+_49741565 | 0.25 |
ENST00000549445.5
ENST00000550951.5 ENST00000549385.5 ENST00000548713.5 ENST00000548201.5 |
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr15_-_82709823 | 0.25 |
ENST00000666973.1
ENST00000664460.1 ENST00000669930.1 |
AP3B2
|
adaptor related protein complex 3 subunit beta 2 |
| chr4_-_147684114 | 0.24 |
ENST00000322396.7
|
PRMT9
|
protein arginine methyltransferase 9 |
| chr15_-_43493076 | 0.24 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr17_+_82830496 | 0.24 |
ENST00000683821.1
|
TBCD
|
tubulin folding cofactor D |
| chr12_+_49741544 | 0.24 |
ENST00000549966.5
ENST00000547832.5 ENST00000547187.5 ENST00000548894.5 ENST00000546914.5 ENST00000552699.5 ENST00000267115.10 |
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr1_+_149832647 | 0.24 |
ENST00000578186.2
|
H4C14
|
H4 clustered histone 14 |
| chr4_-_26490453 | 0.23 |
ENST00000295589.4
|
CCKAR
|
cholecystokinin A receptor |
| chr4_-_185775411 | 0.23 |
ENST00000445115.5
ENST00000451701.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chrX_+_47585212 | 0.23 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr8_-_100106206 | 0.23 |
ENST00000519408.5
|
RGS22
|
regulator of G protein signaling 22 |
| chr2_-_135837170 | 0.23 |
ENST00000264162.7
|
LCT
|
lactase |
| chr3_+_125969172 | 0.22 |
ENST00000514116.6
ENST00000513830.5 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr3_+_98149326 | 0.22 |
ENST00000437310.1
|
OR5H14
|
olfactory receptor family 5 subfamily H member 14 |
| chr19_-_36916021 | 0.21 |
ENST00000520965.5
|
ZNF829
|
zinc finger protein 829 |
| chr2_+_203867764 | 0.21 |
ENST00000648405.2
|
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
| chr9_+_97307645 | 0.21 |
ENST00000529487.3
|
CCDC180
|
coiled-coil domain containing 180 |
| chr3_+_149813806 | 0.21 |
ENST00000468648.5
ENST00000459632.5 |
RNF13
|
ring finger protein 13 |
| chr1_-_60073733 | 0.21 |
ENST00000450089.6
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr3_+_125969152 | 0.20 |
ENST00000251776.8
ENST00000504401.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr3_-_14178569 | 0.20 |
ENST00000285021.12
|
XPC
|
XPC complex subunit, DNA damage recognition and repair factor |
| chr20_+_59676661 | 0.20 |
ENST00000355648.8
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr6_-_27873525 | 0.19 |
ENST00000618305.2
|
H4C13
|
H4 clustered histone 13 |
| chr11_+_35189964 | 0.19 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr21_+_44133610 | 0.19 |
ENST00000644251.1
ENST00000427803.6 ENST00000348499.9 ENST00000291577.11 ENST00000389690.7 |
GATD3A
|
glutamine amidotransferase like class 1 domain containing 3A |
| chr3_-_154430184 | 0.18 |
ENST00000389740.3
|
GPR149
|
G protein-coupled receptor 149 |
| chr1_-_160070125 | 0.18 |
ENST00000639408.1
|
KCNJ10
|
potassium inwardly rectifying channel subfamily J member 10 |
| chr18_+_46174055 | 0.18 |
ENST00000615553.1
|
C18orf25
|
chromosome 18 open reading frame 25 |
| chr16_-_58295019 | 0.17 |
ENST00000567164.6
ENST00000219301.8 ENST00000569727.1 |
PRSS54
|
serine protease 54 |
| chr3_-_112846856 | 0.17 |
ENST00000488794.5
|
CD200R1L
|
CD200 receptor 1 like |
| chrX_-_27981449 | 0.17 |
ENST00000441525.4
|
DCAF8L1
|
DDB1 and CUL4 associated factor 8 like 1 |
| chr3_-_123992046 | 0.17 |
ENST00000467907.5
ENST00000459660.5 ENST00000495093.1 ENST00000460743.5 ENST00000405845.7 ENST00000484329.1 ENST00000479867.1 ENST00000496145.5 |
ROPN1
|
rhophilin associated tail protein 1 |
| chr14_+_91114388 | 0.17 |
ENST00000519019.5
ENST00000523816.5 ENST00000517518.5 |
DGLUCY
|
D-glutamate cyclase |
| chr14_+_91114667 | 0.17 |
ENST00000523894.5
ENST00000522322.5 ENST00000523771.5 |
DGLUCY
|
D-glutamate cyclase |
| chr5_-_180815528 | 0.16 |
ENST00000333055.8
|
MGAT1
|
alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase |
| chr5_-_69332723 | 0.16 |
ENST00000511257.1
ENST00000396496.7 ENST00000383374.6 |
CCDC125
|
coiled-coil domain containing 125 |
| chr6_-_132798587 | 0.16 |
ENST00000275227.9
|
SLC18B1
|
solute carrier family 18 member B1 |
| chr2_+_203867943 | 0.16 |
ENST00000295854.10
ENST00000487393.1 ENST00000472206.1 |
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
| chr8_-_80029904 | 0.15 |
ENST00000521434.5
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr7_-_19773569 | 0.15 |
ENST00000422233.5
ENST00000433641.5 ENST00000405844.6 |
TMEM196
|
transmembrane protein 196 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SMAD3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0060621 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 1.1 | 3.4 | GO:0071613 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 1.0 | 3.0 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.4 | 1.3 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.3 | 0.8 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.2 | 1.0 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.2 | 1.6 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.2 | 1.6 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.2 | 4.8 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.2 | 1.0 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.2 | 0.6 | GO:0035744 | T-helper 1 cell cytokine production(GO:0035744) |
| 0.2 | 1.0 | GO:1990539 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.2 | 0.8 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.2 | 2.3 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 2.5 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.6 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 1.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.8 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 | 1.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.9 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.7 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.1 | 0.9 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 1.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.7 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.3 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.2 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 1.9 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 1.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.4 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.7 | GO:0044821 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.4 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.5 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.7 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.4 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.6 | GO:1903874 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.8 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.8 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.7 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.0 | 0.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.4 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.8 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0070429 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.0 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.4 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.0 | 0.3 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 1.8 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 1.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0044145 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.8 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.8 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 2.5 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.1 | 0.8 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.3 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.3 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 0.5 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 2.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 1.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.0 | 1.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.8 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 1.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 2.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0033265 | choline binding(GO:0033265) |
| 1.0 | 4.8 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.4 | 1.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.3 | 1.0 | GO:0004411 | homogentisate 1,2-dioxygenase activity(GO:0004411) |
| 0.3 | 0.9 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 1.6 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 1.0 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.2 | 2.5 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.2 | 0.8 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.2 | 0.7 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.2 | 1.0 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.8 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.7 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 2.1 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 3.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 3.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.7 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 1.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.5 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.1 | 0.5 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 1.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0030346 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.2 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 2.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 4.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 2.3 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 2.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.9 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |