Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
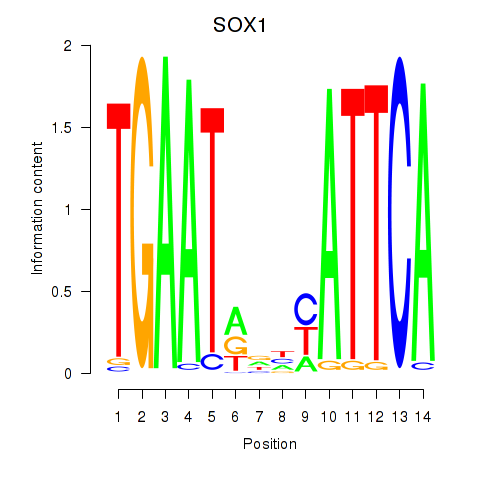
Results for SOX1
Z-value: 0.49
Transcription factors associated with SOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX1
|
ENSG00000182968.5 | SRY-box transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX1 | hg38_v1_chr13_+_112067143_112067160 | 0.09 | 6.4e-01 | Click! |
Activity profile of SOX1 motif
Sorted Z-values of SOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_95069489 | 1.51 |
ENST00000371270.6
ENST00000535898.5 ENST00000623108.3 |
CYP2C8
|
cytochrome P450 family 2 subfamily C member 8 |
| chr16_+_11965193 | 0.89 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr4_+_68815991 | 0.69 |
ENST00000265403.12
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase family 2 member B10 |
| chrX_+_106168297 | 0.68 |
ENST00000337685.6
ENST00000357175.6 |
PWWP3B
|
PWWP domain containing 3B |
| chr19_+_35351552 | 0.68 |
ENST00000246553.3
|
FFAR1
|
free fatty acid receptor 1 |
| chr11_-_47378479 | 0.64 |
ENST00000533968.1
|
SPI1
|
Spi-1 proto-oncogene |
| chr16_+_11965234 | 0.62 |
ENST00000562385.1
|
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr3_-_58214671 | 0.54 |
ENST00000460422.1
ENST00000483681.5 |
DNASE1L3
|
deoxyribonuclease 1 like 3 |
| chr3_-_171771295 | 0.53 |
ENST00000418087.1
|
PLD1
|
phospholipase D1 |
| chr1_-_144456805 | 0.51 |
ENST00000581897.6
ENST00000577412.5 |
NBPF15
|
NBPF member 15 |
| chr11_-_47378391 | 0.50 |
ENST00000227163.8
|
SPI1
|
Spi-1 proto-oncogene |
| chr2_+_188991663 | 0.49 |
ENST00000450867.1
|
COL3A1
|
collagen type III alpha 1 chain |
| chr17_-_31297231 | 0.48 |
ENST00000247271.5
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr1_-_111563956 | 0.46 |
ENST00000369717.8
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr12_-_9999176 | 0.43 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr1_+_32209074 | 0.41 |
ENST00000409358.2
|
DCDC2B
|
doublecortin domain containing 2B |
| chr2_+_101998955 | 0.41 |
ENST00000393414.6
|
IL1R2
|
interleukin 1 receptor type 2 |
| chr17_+_36064265 | 0.38 |
ENST00000616054.2
|
CCL18
|
C-C motif chemokine ligand 18 |
| chr1_-_200409976 | 0.38 |
ENST00000367352.3
|
ZNF281
|
zinc finger protein 281 |
| chr17_-_40665121 | 0.38 |
ENST00000394052.5
|
KRT222
|
keratin 222 |
| chr1_-_12616762 | 0.37 |
ENST00000464917.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr1_-_52033772 | 0.36 |
ENST00000371614.2
|
KTI12
|
KTI12 chromatin associated homolog |
| chr15_-_73753308 | 0.36 |
ENST00000569673.3
|
INSYN1
|
inhibitory synaptic factor 1 |
| chr1_-_111563934 | 0.36 |
ENST00000443498.5
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr15_+_59611776 | 0.34 |
ENST00000396065.3
ENST00000560585.5 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr3_+_11011640 | 0.33 |
ENST00000643396.1
|
SLC6A1
|
solute carrier family 6 member 1 |
| chr1_+_240091866 | 0.33 |
ENST00000319653.14
|
FMN2
|
formin 2 |
| chr1_-_200410001 | 0.32 |
ENST00000367353.2
|
ZNF281
|
zinc finger protein 281 |
| chr6_-_138545685 | 0.32 |
ENST00000342260.9
|
NHSL1
|
NHS like 1 |
| chr19_+_51761167 | 0.31 |
ENST00000340023.7
ENST00000599326.1 ENST00000598953.1 |
FPR2
|
formyl peptide receptor 2 |
| chr6_+_130421086 | 0.30 |
ENST00000545622.5
|
TMEM200A
|
transmembrane protein 200A |
| chr11_-_47378494 | 0.30 |
ENST00000533030.1
|
SPI1
|
Spi-1 proto-oncogene |
| chr1_-_89175997 | 0.30 |
ENST00000294671.3
ENST00000650452.1 |
GBP7
|
guanylate binding protein 7 |
| chr1_+_12746192 | 0.29 |
ENST00000614859.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chr6_+_28281555 | 0.29 |
ENST00000259883.3
ENST00000682144.1 |
PGBD1
|
piggyBac transposable element derived 1 |
| chr1_+_70411241 | 0.28 |
ENST00000370938.8
ENST00000346806.2 |
CTH
|
cystathionine gamma-lyase |
| chr8_-_109680812 | 0.28 |
ENST00000528716.5
ENST00000527600.5 ENST00000531230.5 ENST00000532189.5 ENST00000534184.5 ENST00000408889.7 ENST00000533171.5 |
SYBU
|
syntabulin |
| chr12_-_7695752 | 0.28 |
ENST00000329913.4
|
GDF3
|
growth differentiation factor 3 |
| chr9_+_5890872 | 0.27 |
ENST00000381477.8
ENST00000381476.5 ENST00000381471.1 |
MLANA
|
melan-A |
| chr1_-_183653307 | 0.25 |
ENST00000308641.6
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4 |
| chrX_-_72877864 | 0.25 |
ENST00000596389.5
|
DMRTC1
|
DMRT like family C1 |
| chr3_+_178558700 | 0.25 |
ENST00000432997.5
ENST00000455865.5 |
KCNMB2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr10_-_72217126 | 0.24 |
ENST00000317126.8
|
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr9_-_127847117 | 0.23 |
ENST00000480266.5
|
ENG
|
endoglin |
| chr4_-_170003738 | 0.23 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibril associated protein 3 like |
| chr9_-_101487120 | 0.23 |
ENST00000374848.8
|
PGAP4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr8_-_98942814 | 0.23 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr2_-_162318129 | 0.22 |
ENST00000679938.1
|
IFIH1
|
interferon induced with helicase C domain 1 |
| chr7_+_36410510 | 0.22 |
ENST00000428612.5
|
ANLN
|
anillin actin binding protein |
| chr7_-_106285898 | 0.21 |
ENST00000424768.2
ENST00000681255.1 |
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr8_+_144078590 | 0.20 |
ENST00000525936.1
|
EXOSC4
|
exosome component 4 |
| chr10_-_72216267 | 0.20 |
ENST00000342444.8
ENST00000533958.1 ENST00000672957.1 ENST00000527593.5 ENST00000672940.1 ENST00000530461.5 ENST00000317168.11 ENST00000524829.5 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr3_-_16482850 | 0.20 |
ENST00000432519.5
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr14_+_61697622 | 0.20 |
ENST00000539097.2
|
HIF1A
|
hypoxia inducible factor 1 subunit alpha |
| chr18_+_24139053 | 0.20 |
ENST00000463087.5
ENST00000585037.5 ENST00000399496.8 ENST00000486759.6 ENST00000577705.1 ENST00000415309.6 ENST00000621648.4 ENST00000581397.5 |
CABYR
|
calcium binding tyrosine phosphorylation regulated |
| chr1_+_184051678 | 0.19 |
ENST00000643231.1
|
TSEN15
|
tRNA splicing endonuclease subunit 15 |
| chr9_-_13165442 | 0.19 |
ENST00000542239.1
ENST00000538841.5 ENST00000433359.6 |
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr3_+_138608851 | 0.18 |
ENST00000393035.6
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_-_200410052 | 0.18 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr8_+_144078661 | 0.17 |
ENST00000316052.6
|
EXOSC4
|
exosome component 4 |
| chr3_-_190122317 | 0.17 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr20_+_50958805 | 0.17 |
ENST00000244051.3
|
MOCS3
|
molybdenum cofactor synthesis 3 |
| chr7_-_105389365 | 0.16 |
ENST00000482897.5
|
SRPK2
|
SRSF protein kinase 2 |
| chr1_+_174799895 | 0.16 |
ENST00000489615.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr3_+_126394890 | 0.16 |
ENST00000352312.6
|
CFAP100
|
cilia and flagella associated protein 100 |
| chr3_+_128051610 | 0.15 |
ENST00000464451.5
|
SEC61A1
|
SEC61 translocon subunit alpha 1 |
| chr8_-_109608055 | 0.15 |
ENST00000529690.5
|
SYBU
|
syntabulin |
| chr2_-_199456046 | 0.15 |
ENST00000428695.5
|
SATB2
|
SATB homeobox 2 |
| chr2_+_209580024 | 0.15 |
ENST00000392194.5
|
MAP2
|
microtubule associated protein 2 |
| chr22_-_30266839 | 0.15 |
ENST00000403463.1
ENST00000215781.3 |
OSM
|
oncostatin M |
| chr1_+_65147657 | 0.15 |
ENST00000546702.5
|
AK4
|
adenylate kinase 4 |
| chr1_+_65147514 | 0.14 |
ENST00000545314.5
|
AK4
|
adenylate kinase 4 |
| chr2_-_208190001 | 0.14 |
ENST00000451346.5
ENST00000341287.9 |
C2orf80
|
chromosome 2 open reading frame 80 |
| chr1_-_86383078 | 0.14 |
ENST00000460698.6
|
ODF2L
|
outer dense fiber of sperm tails 2 like |
| chr11_-_113706292 | 0.13 |
ENST00000544634.5
ENST00000539732.5 ENST00000538770.1 ENST00000536856.5 ENST00000299882.11 ENST00000544476.1 |
TMPRSS5
|
transmembrane serine protease 5 |
| chr19_-_6057271 | 0.13 |
ENST00000592281.5
|
RFX2
|
regulatory factor X2 |
| chr14_+_22501542 | 0.13 |
ENST00000390498.1
|
TRAJ39
|
T cell receptor alpha joining 39 |
| chr1_+_184051696 | 0.12 |
ENST00000533373.6
ENST00000647437.1 ENST00000647465.1 ENST00000645668.2 ENST00000645963.2 |
TSEN15
|
tRNA splicing endonuclease subunit 15 |
| chr13_+_108596152 | 0.12 |
ENST00000356711.7
ENST00000251041.10 |
MYO16
|
myosin XVI |
| chr3_+_138608700 | 0.12 |
ENST00000360570.7
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_-_120325936 | 0.12 |
ENST00000549767.1
|
PLA2G1B
|
phospholipase A2 group IB |
| chr17_+_82858519 | 0.12 |
ENST00000576160.6
ENST00000571712.5 |
TBCD
|
tubulin folding cofactor D |
| chr1_+_184051643 | 0.12 |
ENST00000423085.7
ENST00000361641.6 |
TSEN15
|
tRNA splicing endonuclease subunit 15 |
| chr5_-_55692731 | 0.11 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38 member 9 |
| chr17_-_81166160 | 0.11 |
ENST00000326724.9
|
AATK
|
apoptosis associated tyrosine kinase |
| chrX_-_63755032 | 0.10 |
ENST00000624538.2
ENST00000636276.1 ENST00000624843.3 ENST00000671907.1 ENST00000624210.3 ENST00000374870.8 ENST00000635967.1 ENST00000253401.10 ENST00000672194.1 ENST00000637723.2 ENST00000637417.1 ENST00000637520.1 ENST00000374872.4 ENST00000636926.1 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chr1_-_43172504 | 0.10 |
ENST00000431635.6
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr19_+_52397841 | 0.10 |
ENST00000360465.8
ENST00000391788.6 ENST00000436397.5 ENST00000391787.6 ENST00000494167.6 ENST00000493272.5 |
ZNF528
|
zinc finger protein 528 |
| chr13_-_21604145 | 0.10 |
ENST00000382374.9
ENST00000468222.2 |
MICU2
|
mitochondrial calcium uptake 2 |
| chr11_-_113706330 | 0.10 |
ENST00000540540.5
ENST00000538955.5 ENST00000545579.6 |
TMPRSS5
|
transmembrane serine protease 5 |
| chr2_-_162318475 | 0.09 |
ENST00000648433.1
|
IFIH1
|
interferon induced with helicase C domain 1 |
| chr2_-_96260575 | 0.09 |
ENST00000435268.1
|
TMEM127
|
transmembrane protein 127 |
| chr3_+_138608575 | 0.09 |
ENST00000338446.8
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr11_-_57410113 | 0.08 |
ENST00000529411.1
|
ENSG00000254979.5
|
novel protein |
| chrX_-_63755187 | 0.08 |
ENST00000635729.1
ENST00000623566.3 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chr2_-_208189985 | 0.08 |
ENST00000449053.1
|
C2orf80
|
chromosome 2 open reading frame 80 |
| chr8_-_133489631 | 0.08 |
ENST00000523634.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr22_+_41952132 | 0.08 |
ENST00000381348.4
|
LINC00634
|
long intergenic non-protein coding RNA 634 |
| chr11_-_134253248 | 0.08 |
ENST00000392595.6
ENST00000352327.5 ENST00000341541.8 ENST00000392594.7 |
THYN1
|
thymocyte nuclear protein 1 |
| chr7_+_142760398 | 0.07 |
ENST00000632998.1
|
PRSS2
|
serine protease 2 |
| chr19_+_54874218 | 0.07 |
ENST00000355524.8
ENST00000391726.7 |
FCAR
|
Fc fragment of IgA receptor |
| chr11_-_112260860 | 0.07 |
ENST00000338832.4
|
PLET1
|
placenta expressed transcript 1 |
| chr19_+_3762665 | 0.06 |
ENST00000330133.5
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr1_-_19251509 | 0.06 |
ENST00000375199.7
ENST00000375208.7 ENST00000477853.6 |
EMC1
|
ER membrane protein complex subunit 1 |
| chr3_+_158644510 | 0.06 |
ENST00000486715.6
ENST00000478576.5 ENST00000264263.9 ENST00000464732.1 |
GFM1
|
G elongation factor mitochondrial 1 |
| chr7_-_26200734 | 0.06 |
ENST00000354667.8
ENST00000618183.5 |
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr11_-_76669985 | 0.06 |
ENST00000407242.6
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr13_+_32316063 | 0.06 |
ENST00000680887.1
|
BRCA2
|
BRCA2 DNA repair associated |
| chr6_-_130222813 | 0.06 |
ENST00000437477.6
ENST00000439090.7 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr19_+_38389612 | 0.05 |
ENST00000586301.5
|
SPRED3
|
sprouty related EVH1 domain containing 3 |
| chr2_-_162318613 | 0.05 |
ENST00000649979.2
ENST00000421365.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr21_-_30561314 | 0.04 |
ENST00000334849.2
|
KRTAP19-7
|
keratin associated protein 19-7 |
| chr9_-_104536856 | 0.04 |
ENST00000641090.1
|
OR13C3
|
olfactory receptor family 13 subfamily C member 3 |
| chrX_-_116462965 | 0.04 |
ENST00000371894.5
|
CT83
|
cancer/testis antigen 83 |
| chr12_+_7304284 | 0.04 |
ENST00000399422.4
|
ACSM4
|
acyl-CoA synthetase medium chain family member 4 |
| chr1_-_53940100 | 0.04 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr2_+_170178136 | 0.04 |
ENST00000409044.7
ENST00000408978.9 |
MYO3B
|
myosin IIIB |
| chr8_-_92017292 | 0.04 |
ENST00000521553.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr6_+_123804141 | 0.04 |
ENST00000368416.5
|
NKAIN2
|
sodium/potassium transporting ATPase interacting 2 |
| chr14_+_22040576 | 0.03 |
ENST00000390448.3
|
TRAV20
|
T cell receptor alpha variable 20 |
| chr12_-_64752871 | 0.03 |
ENST00000418919.6
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chrX_-_120878924 | 0.03 |
ENST00000613352.1
|
ENSG00000278646.1
|
novel protein similar to cancer/testis antigen family 47, member A12 (CT47A12) |
| chr15_+_42273194 | 0.03 |
ENST00000561871.1
|
GANC
|
glucosidase alpha, neutral C |
| chr11_-_33753394 | 0.03 |
ENST00000532057.5
ENST00000531080.5 |
FBXO3
|
F-box protein 3 |
| chr11_+_5422111 | 0.03 |
ENST00000300778.4
|
OR51Q1
|
olfactory receptor family 51 subfamily Q member 1 |
| chr3_-_44477639 | 0.03 |
ENST00000396077.8
|
ZNF445
|
zinc finger protein 445 |
| chr17_+_76732978 | 0.03 |
ENST00000587459.1
|
ENSG00000267168.1
|
novel protein |
| chr2_+_201116396 | 0.02 |
ENST00000395148.6
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr11_-_133845495 | 0.01 |
ENST00000299140.8
ENST00000532889.1 |
SPATA19
|
spermatogenesis associated 19 |
| chr18_-_59318544 | 0.01 |
ENST00000587244.5
|
CPLX4
|
complexin 4 |
| chr10_-_13972355 | 0.01 |
ENST00000264546.10
|
FRMD4A
|
FERM domain containing 4A |
| chr4_-_163344832 | 0.01 |
ENST00000511901.1
|
NPY1R
|
neuropeptide Y receptor Y1 |
| chr11_-_124313949 | 0.01 |
ENST00000641015.1
|
OR8D1
|
olfactory receptor family 8 subfamily D member 1 |
| chr11_-_76670737 | 0.01 |
ENST00000260061.9
ENST00000404995.5 |
LRRC32
|
leucine rich repeat containing 32 |
| chrX_+_22032427 | 0.00 |
ENST00000684143.1
|
PHEX
|
phosphate regulating endopeptidase homolog X-linked |
| chr8_-_85378105 | 0.00 |
ENST00000521846.5
ENST00000523022.6 ENST00000524324.5 ENST00000519991.5 ENST00000520663.5 ENST00000517590.5 ENST00000522579.5 ENST00000522814.5 ENST00000522662.5 ENST00000523858.5 ENST00000519129.5 |
CA1
|
carbonic anhydrase 1 |
| chr9_-_111328520 | 0.00 |
ENST00000374428.1
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2 |
| chr6_-_49969625 | 0.00 |
ENST00000398718.1
|
DEFB113
|
defensin beta 113 |
| chr11_+_6863057 | 0.00 |
ENST00000641461.1
|
OR10A2
|
olfactory receptor family 10 subfamily A member 2 |
| chr6_+_49499965 | 0.00 |
ENST00000545705.1
|
GLYATL3
|
glycine-N-acyltransferase like 3 |
| chr6_+_49499916 | 0.00 |
ENST00000371197.9
|
GLYATL3
|
glycine-N-acyltransferase like 3 |
| chr11_+_125626724 | 0.00 |
ENST00000531607.5
|
CHEK1
|
checkpoint kinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.2 | 1.4 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.3 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 0.1 | 0.4 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.3 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.2 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.1 | 0.4 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.3 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.4 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.3 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.2 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.1 | 0.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.2 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.0 | 0.5 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.3 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.5 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.3 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.2 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.5 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.8 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.2 | GO:0097229 | sperm end piece(GO:0097229) |
| 0.0 | 0.2 | GO:0097545 | axonemal outer doublet(GO:0097545) |
| 0.0 | 0.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0033593 | BRCA2-MAGE-D1 complex(GO:0033593) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.3 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 1.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.4 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.3 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.7 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |