Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
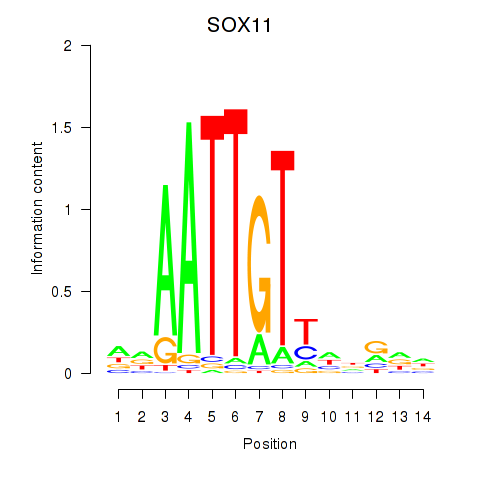
Results for SOX11
Z-value: 0.99
Transcription factors associated with SOX11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX11
|
ENSG00000176887.7 | SRY-box transcription factor 11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX11 | hg38_v1_chr2_+_5692357_5692410 | 0.06 | 7.2e-01 | Click! |
Activity profile of SOX11 motif
Sorted Z-values of SOX11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_68955332 | 3.46 |
ENST00000269080.6
ENST00000615593.4 ENST00000586539.6 ENST00000430352.6 |
ABCA8
|
ATP binding cassette subfamily A member 8 |
| chrX_-_21758097 | 3.28 |
ENST00000379494.4
|
SMPX
|
small muscle protein X-linked |
| chrX_-_21758021 | 2.91 |
ENST00000646008.1
|
SMPX
|
small muscle protein X-linked |
| chr5_+_72107453 | 2.83 |
ENST00000296755.12
ENST00000511641.2 |
MAP1B
|
microtubule associated protein 1B |
| chr2_+_168802610 | 2.76 |
ENST00000397206.6
ENST00000317647.12 ENST00000397209.6 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr6_-_46080332 | 2.63 |
ENST00000185206.12
|
CLIC5
|
chloride intracellular channel 5 |
| chrX_-_15600953 | 2.50 |
ENST00000679212.1
ENST00000679278.1 ENST00000678046.1 ENST00000252519.8 |
ACE2
|
angiotensin I converting enzyme 2 |
| chr2_+_168802563 | 2.15 |
ENST00000445023.6
|
NOSTRIN
|
nitric oxide synthase trafficking |
| chr13_-_110242694 | 2.12 |
ENST00000648989.1
ENST00000647797.1 ENST00000648966.1 ENST00000649484.1 ENST00000648695.1 ENST00000650115.1 ENST00000650566.1 |
COL4A1
|
collagen type IV alpha 1 chain |
| chr10_+_80408485 | 2.07 |
ENST00000615554.4
ENST00000372185.5 |
PRXL2A
|
peroxiredoxin like 2A |
| chrX_-_13817027 | 1.97 |
ENST00000493677.5
ENST00000355135.6 ENST00000316715.9 |
GPM6B
|
glycoprotein M6B |
| chr2_+_69013414 | 1.95 |
ENST00000681816.1
ENST00000482235.2 |
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr11_-_118176576 | 1.88 |
ENST00000278947.6
|
SCN2B
|
sodium voltage-gated channel beta subunit 2 |
| chrM_+_8366 | 1.78 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase membrane subunit 8 |
| chr6_-_75363003 | 1.77 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr12_-_21910853 | 1.74 |
ENST00000544039.5
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr2_+_69013379 | 1.67 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr9_+_12775012 | 1.65 |
ENST00000319264.4
|
LURAP1L
|
leucine rich adaptor protein 1 like |
| chr6_+_54018910 | 1.64 |
ENST00000514921.5
ENST00000274897.9 ENST00000370877.6 |
MLIP
|
muscular LMNA interacting protein |
| chr9_-_92404559 | 1.62 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chrM_+_8489 | 1.59 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase membrane subunit 6 |
| chr18_+_7754959 | 1.56 |
ENST00000400053.8
|
PTPRM
|
protein tyrosine phosphatase receptor type M |
| chr7_+_80624071 | 1.56 |
ENST00000438020.5
|
CD36
|
CD36 molecule |
| chrM_+_7586 | 1.54 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr8_-_13276491 | 1.53 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr2_+_102311546 | 1.51 |
ENST00000233954.6
ENST00000447231.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr4_-_185649524 | 1.46 |
ENST00000451974.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr8_-_85341705 | 1.41 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr5_+_149141890 | 1.41 |
ENST00000508983.5
|
ABLIM3
|
actin binding LIM protein family member 3 |
| chr7_-_7535863 | 1.37 |
ENST00000399429.8
|
COL28A1
|
collagen type XXVIII alpha 1 chain |
| chr5_+_149141817 | 1.34 |
ENST00000504238.5
|
ABLIM3
|
actin binding LIM protein family member 3 |
| chr8_-_85341659 | 1.32 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr13_-_44474296 | 1.29 |
ENST00000611198.4
|
TSC22D1
|
TSC22 domain family member 1 |
| chr2_-_224402097 | 1.26 |
ENST00000409685.4
|
FAM124B
|
family with sequence similarity 124 member B |
| chr12_-_106084023 | 1.26 |
ENST00000553094.1
ENST00000549704.1 |
NUAK1
|
NUAK family kinase 1 |
| chr1_+_205042960 | 1.22 |
ENST00000638378.1
|
CNTN2
|
contactin 2 |
| chr2_-_224402060 | 1.18 |
ENST00000243806.2
|
FAM124B
|
family with sequence similarity 124 member B |
| chr2_-_224401994 | 1.17 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124 member B |
| chr1_+_65264694 | 1.11 |
ENST00000263441.11
ENST00000395325.7 |
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr5_+_167284799 | 1.10 |
ENST00000518659.5
|
TENM2
|
teneurin transmembrane protein 2 |
| chrX_-_32155462 | 1.10 |
ENST00000359836.5
ENST00000378707.7 ENST00000541735.5 ENST00000684130.1 ENST00000682238.1 ENST00000620040.5 ENST00000474231.5 |
DMD
|
dystrophin |
| chr15_-_99251207 | 1.05 |
ENST00000394129.6
ENST00000558663.5 ENST00000394135.7 ENST00000561365.5 ENST00000560279.5 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr15_+_80441229 | 1.04 |
ENST00000533983.5
ENST00000527771.5 ENST00000525103.1 |
ARNT2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr4_-_89838289 | 1.04 |
ENST00000336904.7
|
SNCA
|
synuclein alpha |
| chr7_-_93890160 | 1.03 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr20_+_11892493 | 1.03 |
ENST00000422390.5
ENST00000618918.4 |
BTBD3
|
BTB domain containing 3 |
| chr2_+_102311502 | 0.99 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr1_+_204870831 | 0.98 |
ENST00000404076.5
ENST00000539706.6 |
NFASC
|
neurofascin |
| chr17_-_36001549 | 0.98 |
ENST00000617897.2
|
CCL15
|
C-C motif chemokine ligand 15 |
| chr15_+_83447328 | 0.96 |
ENST00000427482.7
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3 |
| chr16_+_482507 | 0.96 |
ENST00000412256.1
|
RAB11FIP3
|
RAB11 family interacting protein 3 |
| chr5_+_174045673 | 0.95 |
ENST00000303177.8
ENST00000519867.5 |
NSG2
|
neuronal vesicle trafficking associated 2 |
| chr15_-_79090760 | 0.95 |
ENST00000419573.7
ENST00000558480.7 |
RASGRF1
|
Ras protein specific guanine nucleotide releasing factor 1 |
| chr6_-_47309898 | 0.94 |
ENST00000296861.2
|
TNFRSF21
|
TNF receptor superfamily member 21 |
| chr22_-_33572227 | 0.94 |
ENST00000674780.1
|
LARGE1
|
LARGE xylosyl- and glucuronyltransferase 1 |
| chr8_+_104339796 | 0.94 |
ENST00000622554.1
ENST00000297581.2 |
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr6_+_30617825 | 0.94 |
ENST00000259873.5
|
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr14_-_52069039 | 0.90 |
ENST00000216286.10
|
NID2
|
nidogen 2 |
| chr8_+_24384275 | 0.88 |
ENST00000256412.8
|
ADAMDEC1
|
ADAM like decysin 1 |
| chr4_+_143391506 | 0.87 |
ENST00000509992.1
|
GAB1
|
GRB2 associated binding protein 1 |
| chr17_-_10114546 | 0.85 |
ENST00000323816.8
|
GAS7
|
growth arrest specific 7 |
| chr10_+_68109433 | 0.85 |
ENST00000613327.4
ENST00000358913.10 ENST00000373675.3 |
MYPN
|
myopalladin |
| chr6_+_54018992 | 0.84 |
ENST00000509997.5
|
MLIP
|
muscular LMNA interacting protein |
| chr4_+_158806069 | 0.84 |
ENST00000512986.5
|
FNIP2
|
folliculin interacting protein 2 |
| chr12_-_15712910 | 0.84 |
ENST00000543612.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr4_+_71339014 | 0.82 |
ENST00000340595.4
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr13_+_75839716 | 0.80 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr18_+_48539017 | 0.80 |
ENST00000256413.8
|
CTIF
|
cap binding complex dependent translation initiation factor |
| chr6_-_49636832 | 0.79 |
ENST00000371175.10
ENST00000646272.1 ENST00000646939.1 ENST00000618248.3 ENST00000229810.9 ENST00000646963.1 |
RHAG
|
Rh associated glycoprotein |
| chr12_+_59664677 | 0.76 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr10_+_68106109 | 0.75 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr18_+_48539112 | 0.75 |
ENST00000382998.8
|
CTIF
|
cap binding complex dependent translation initiation factor |
| chr2_+_33458043 | 0.73 |
ENST00000437184.5
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr6_-_11382247 | 0.72 |
ENST00000397378.7
ENST00000513989.5 ENST00000508546.5 ENST00000504387.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr7_-_143882808 | 0.72 |
ENST00000460532.5
ENST00000491908.1 |
TCAF1
|
TRPM8 channel associated factor 1 |
| chr6_-_56628018 | 0.72 |
ENST00000522360.5
|
DST
|
dystonin |
| chr1_-_112935984 | 0.71 |
ENST00000443580.6
|
SLC16A1
|
solute carrier family 16 member 1 |
| chr18_+_48539198 | 0.71 |
ENST00000591412.5
|
CTIF
|
cap binding complex dependent translation initiation factor |
| chr1_-_247724043 | 0.71 |
ENST00000366485.1
|
OR14A2
|
olfactory receptor family 14 subfamily A member 2 |
| chr1_-_158399466 | 0.71 |
ENST00000334438.1
|
OR10T2
|
olfactory receptor family 10 subfamily T member 2 |
| chr16_+_19067893 | 0.70 |
ENST00000544894.6
ENST00000561858.5 |
COQ7
|
coenzyme Q7, hydroxylase |
| chr4_-_22443110 | 0.69 |
ENST00000508133.5
|
ADGRA3
|
adhesion G protein-coupled receptor A3 |
| chr9_-_14300231 | 0.68 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B |
| chr15_+_83447411 | 0.68 |
ENST00000324537.5
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3 |
| chrX_-_15601077 | 0.68 |
ENST00000680121.1
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr6_+_148508446 | 0.63 |
ENST00000637729.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr9_+_27109135 | 0.63 |
ENST00000519097.5
ENST00000615002.4 |
TEK
|
TEK receptor tyrosine kinase |
| chr4_+_140373474 | 0.62 |
ENST00000512749.5
ENST00000506597.2 ENST00000608372.6 ENST00000510586.5 |
SCOC
|
short coiled-coil protein |
| chr18_+_59220149 | 0.62 |
ENST00000256857.7
ENST00000529320.2 ENST00000420468.6 |
GRP
|
gastrin releasing peptide |
| chr19_-_5680488 | 0.61 |
ENST00000587589.1
ENST00000309324.9 |
MICOS13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chr7_-_83162899 | 0.60 |
ENST00000423517.6
|
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr8_-_71361860 | 0.60 |
ENST00000303824.11
ENST00000645451.1 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr8_-_71362054 | 0.59 |
ENST00000340726.8
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr11_+_22666604 | 0.55 |
ENST00000454584.6
|
GAS2
|
growth arrest specific 2 |
| chr15_+_98890341 | 0.55 |
ENST00000558898.1
|
IGF1R
|
insulin like growth factor 1 receptor |
| chr4_-_46388713 | 0.55 |
ENST00000507069.5
|
GABRA2
|
gamma-aminobutyric acid type A receptor subunit alpha2 |
| chr4_+_71339037 | 0.54 |
ENST00000512686.5
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr12_+_40827992 | 0.51 |
ENST00000547849.5
|
CNTN1
|
contactin 1 |
| chr2_-_189179754 | 0.50 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr21_+_42199686 | 0.50 |
ENST00000398457.6
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr1_-_110519175 | 0.49 |
ENST00000369771.4
|
KCNA10
|
potassium voltage-gated channel subfamily A member 10 |
| chr2_-_110212519 | 0.49 |
ENST00000611969.5
|
MTLN
|
mitoregulin |
| chr2_-_2324323 | 0.49 |
ENST00000648339.1
ENST00000647694.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr4_+_87832917 | 0.48 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chrX_-_133097095 | 0.48 |
ENST00000511190.5
|
USP26
|
ubiquitin specific peptidase 26 |
| chr1_+_152970646 | 0.47 |
ENST00000328051.3
|
SPRR4
|
small proline rich protein 4 |
| chr4_-_108762535 | 0.45 |
ENST00000512320.1
ENST00000510723.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr7_-_83162857 | 0.44 |
ENST00000333891.14
|
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr1_-_247723764 | 0.44 |
ENST00000419891.1
|
ENSG00000239395.2
|
novel protein |
| chr9_+_27109200 | 0.43 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase |
| chr1_-_145910031 | 0.43 |
ENST00000369304.8
|
ITGA10
|
integrin subunit alpha 10 |
| chr6_+_26183750 | 0.42 |
ENST00000614097.3
|
H2BC6
|
H2B clustered histone 6 |
| chr13_-_75366973 | 0.40 |
ENST00000648194.1
|
TBC1D4
|
TBC1 domain family member 4 |
| chr13_-_44474250 | 0.38 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family member 1 |
| chr5_+_108747879 | 0.38 |
ENST00000281092.9
|
FER
|
FER tyrosine kinase |
| chr9_-_21368962 | 0.38 |
ENST00000610660.1
|
IFNA13
|
interferon alpha 13 |
| chr18_-_72865680 | 0.38 |
ENST00000397929.5
|
NETO1
|
neuropilin and tolloid like 1 |
| chr3_+_184032831 | 0.38 |
ENST00000382489.3
|
HTR3D
|
5-hydroxytryptamine receptor 3D |
| chr14_+_96482982 | 0.37 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr6_-_48111132 | 0.36 |
ENST00000398738.3
ENST00000679966.1 ENST00000339488.9 |
PTCHD4
|
patched domain containing 4 |
| chr1_-_153150884 | 0.36 |
ENST00000368748.5
|
SPRR2G
|
small proline rich protein 2G |
| chr1_-_145910066 | 0.35 |
ENST00000539363.2
|
ITGA10
|
integrin subunit alpha 10 |
| chr13_-_98977975 | 0.35 |
ENST00000376460.5
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr6_-_87291988 | 0.34 |
ENST00000369576.2
|
GJB7
|
gap junction protein beta 7 |
| chrX_-_7927701 | 0.34 |
ENST00000537427.5
ENST00000444736.5 ENST00000442940.1 |
PNPLA4
|
patatin like phospholipase domain containing 4 |
| chr5_-_116536458 | 0.34 |
ENST00000510263.5
|
SEMA6A
|
semaphorin 6A |
| chr12_+_55681711 | 0.33 |
ENST00000394252.4
|
METTL7B
|
methyltransferase like 7B |
| chr14_-_20333268 | 0.32 |
ENST00000358932.9
ENST00000557665.5 |
CCNB1IP1
|
cyclin B1 interacting protein 1 |
| chr4_-_104494882 | 0.32 |
ENST00000394767.3
|
CXXC4
|
CXXC finger protein 4 |
| chr16_+_19067989 | 0.31 |
ENST00000569127.1
|
COQ7
|
coenzyme Q7, hydroxylase |
| chr7_-_93226449 | 0.31 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr11_-_58844484 | 0.31 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase like 2 |
| chr6_-_31158073 | 0.30 |
ENST00000507751.5
ENST00000448162.6 ENST00000502557.5 ENST00000503420.5 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.5 ENST00000396263.6 ENST00000508683.5 ENST00000428174.1 ENST00000448141.6 ENST00000507829.5 ENST00000455279.6 ENST00000376266.9 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr18_+_58196736 | 0.30 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_-_114749900 | 0.30 |
ENST00000438362.6
|
CSDE1
|
cold shock domain containing E1 |
| chr8_+_9555900 | 0.29 |
ENST00000310430.11
ENST00000520408.5 ENST00000522110.1 |
TNKS
|
tankyrase |
| chr12_+_40828173 | 0.29 |
ENST00000552913.5
|
CNTN1
|
contactin 1 |
| chr9_+_104504263 | 0.29 |
ENST00000334726.3
|
OR13F1
|
olfactory receptor family 13 subfamily F member 1 |
| chr12_-_10802627 | 0.29 |
ENST00000240687.2
|
TAS2R7
|
taste 2 receptor member 7 |
| chr9_-_21141832 | 0.29 |
ENST00000380229.4
|
IFNW1
|
interferon omega 1 |
| chr8_-_53839846 | 0.27 |
ENST00000520188.5
|
ATP6V1H
|
ATPase H+ transporting V1 subunit H |
| chr14_-_20333306 | 0.27 |
ENST00000353689.8
ENST00000437553.6 |
CCNB1IP1
|
cyclin B1 interacting protein 1 |
| chr3_-_161346295 | 0.26 |
ENST00000620149.1
|
SPTSSB
|
serine palmitoyltransferase small subunit B |
| chr5_-_116536428 | 0.25 |
ENST00000515009.5
|
SEMA6A
|
semaphorin 6A |
| chr11_-_58844695 | 0.25 |
ENST00000287275.6
|
GLYATL2
|
glycine-N-acyltransferase like 2 |
| chr2_+_165469647 | 0.25 |
ENST00000421875.5
ENST00000314499.11 ENST00000409664.5 ENST00000651982.1 |
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr12_-_81369348 | 0.25 |
ENST00000548670.1
ENST00000541017.5 ENST00000541570.6 ENST00000553058.5 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr17_+_37375974 | 0.24 |
ENST00000615133.2
ENST00000611038.4 |
C17orf78
|
chromosome 17 open reading frame 78 |
| chr5_+_174045741 | 0.23 |
ENST00000521585.5
|
NSG2
|
neuronal vesicle trafficking associated 2 |
| chr8_-_81461574 | 0.23 |
ENST00000379071.4
|
FABP9
|
fatty acid binding protein 9 |
| chr10_-_13890643 | 0.23 |
ENST00000649246.1
|
FRMD4A
|
FERM domain containing 4A |
| chr21_+_5011799 | 0.22 |
ENST00000624081.1
|
ENSG00000279493.1
|
novel protein, similar to DNA (cytosine-5-)-methyltransferase 3-like DNMT3L |
| chr12_+_55681647 | 0.22 |
ENST00000614691.1
|
METTL7B
|
methyltransferase like 7B |
| chr20_+_33237712 | 0.20 |
ENST00000618484.1
|
BPIFA1
|
BPI fold containing family A member 1 |
| chr5_+_140868945 | 0.20 |
ENST00000398640.7
|
PCDHA11
|
protocadherin alpha 11 |
| chrY_+_22973819 | 0.20 |
ENST00000602732.5
|
BPY2
|
basic charge Y-linked 2 |
| chr1_+_196652022 | 0.19 |
ENST00000367429.9
ENST00000630130.2 ENST00000359637.2 |
CFH
|
complement factor H |
| chr1_+_81699665 | 0.18 |
ENST00000359929.7
|
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr3_-_46085817 | 0.17 |
ENST00000683768.1
|
XCR1
|
X-C motif chemokine receptor 1 |
| chr5_-_55173173 | 0.16 |
ENST00000296733.5
ENST00000322374.10 ENST00000381375.7 |
CDC20B
|
cell division cycle 20B |
| chr6_-_116060859 | 0.16 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr5_+_140827950 | 0.16 |
ENST00000378126.4
ENST00000529310.6 ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr5_+_140834230 | 0.16 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr4_+_158521714 | 0.16 |
ENST00000613319.4
ENST00000423548.5 ENST00000448688.6 |
RXFP1
|
relaxin family peptide receptor 1 |
| chr12_+_40742342 | 0.15 |
ENST00000548005.5
ENST00000552248.5 |
CNTN1
|
contactin 1 |
| chr7_+_99325857 | 0.15 |
ENST00000638617.1
ENST00000262942.10 |
ENSG00000284292.1
ARPC1A
|
novel protein, ARPC1A and ARPC1B readthrough actin related protein 2/3 complex subunit 1A |
| chr6_+_49499965 | 0.15 |
ENST00000545705.1
|
GLYATL3
|
glycine-N-acyltransferase like 3 |
| chr4_+_158521937 | 0.15 |
ENST00000343542.9
ENST00000470033.2 |
RXFP1
|
relaxin family peptide receptor 1 |
| chr9_+_127264740 | 0.13 |
ENST00000373387.9
|
GARNL3
|
GTPase activating Rap/RanGAP domain like 3 |
| chr18_+_24460655 | 0.13 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr10_+_70093626 | 0.13 |
ENST00000678195.1
|
MACROH2A2
|
macroH2A.2 histone |
| chr7_+_70596078 | 0.13 |
ENST00000644506.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr3_-_167474026 | 0.13 |
ENST00000466903.1
ENST00000264677.8 |
SERPINI2
|
serpin family I member 2 |
| chr5_-_160852200 | 0.12 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr10_-_125816596 | 0.11 |
ENST00000368786.5
|
UROS
|
uroporphyrinogen III synthase |
| chr7_+_144095048 | 0.11 |
ENST00000408949.2
|
OR2A12
|
olfactory receptor family 2 subfamily A member 12 |
| chrX_+_27807990 | 0.11 |
ENST00000356790.2
|
MAGEB10
|
MAGE family member B10 |
| chr11_-_46826842 | 0.11 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr4_+_70154220 | 0.10 |
ENST00000344526.10
|
PRR27
|
proline rich 27 |
| chr7_-_81770013 | 0.10 |
ENST00000465234.2
|
HGF
|
hepatocyte growth factor |
| chrX_+_26138343 | 0.10 |
ENST00000325250.2
|
MAGEB18
|
MAGE family member B18 |
| chr14_+_22524325 | 0.09 |
ENST00000390517.1
|
TRAJ20
|
T cell receptor alpha joining 20 |
| chr2_+_137964279 | 0.09 |
ENST00000329366.8
|
HNMT
|
histamine N-methyltransferase |
| chr5_+_170353480 | 0.09 |
ENST00000377360.8
|
KCNIP1
|
potassium voltage-gated channel interacting protein 1 |
| chr7_+_142111739 | 0.09 |
ENST00000550469.6
ENST00000477922.3 |
MGAM2
|
maltase-glucoamylase 2 (putative) |
| chr11_+_59878809 | 0.08 |
ENST00000646438.1
|
OOSP3
|
oocyte secreted protein family member 3 |
| chr6_+_52423680 | 0.08 |
ENST00000538167.2
|
EFHC1
|
EF-hand domain containing 1 |
| chr17_+_6444441 | 0.07 |
ENST00000250056.12
ENST00000571373.5 ENST00000570337.6 ENST00000572595.6 ENST00000572447.6 ENST00000576056.5 |
PIMREG
|
PICALM interacting mitotic regulator |
| chr6_+_29100609 | 0.07 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor family 2 subfamily J member 1 |
| chr3_+_178536205 | 0.07 |
ENST00000420517.6
|
KCNMB2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr15_+_36702009 | 0.07 |
ENST00000562489.1
|
CDIN1
|
CDAN1 interacting nuclease 1 |
| chr18_+_24460630 | 0.06 |
ENST00000256906.5
|
HRH4
|
histamine receptor H4 |
| chr12_-_10807286 | 0.05 |
ENST00000240615.3
|
TAS2R8
|
taste 2 receptor member 8 |
| chr8_+_117135259 | 0.05 |
ENST00000519688.5
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr11_-_123977781 | 0.05 |
ENST00000641123.1
|
OR10S1
|
olfactory receptor family 10 subfamily S member 1 |
| chr22_-_28306645 | 0.04 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr5_+_55853314 | 0.04 |
ENST00000354961.8
ENST00000297015.7 |
IL31RA
|
interleukin 31 receptor A |
| chr4_-_106368772 | 0.04 |
ENST00000638719.4
|
GIMD1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr4_+_158521872 | 0.03 |
ENST00000307765.10
|
RXFP1
|
relaxin family peptide receptor 1 |
| chr11_-_5778667 | 0.03 |
ENST00000317093.2
|
OR52N5
|
olfactory receptor family 52 subfamily N member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.6 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.6 | 3.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.5 | 3.2 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.4 | 2.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.4 | 1.2 | GO:0071206 | establishment of protein localization to juxtaparanode region of axon(GO:0071206) |
| 0.3 | 1.0 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 2.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 0.8 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.2 | 0.9 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.2 | 1.9 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 3.0 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.9 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) osteoclast fusion(GO:0072675) |
| 0.2 | 3.5 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.2 | 0.5 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.1 | 1.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.6 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 1.6 | GO:2000332 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 1.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 1.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 2.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 2.8 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.1 | 1.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.3 | GO:0034092 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.1 | 0.4 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.6 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 1.5 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 1.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.7 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 1.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.5 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.1 | 1.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 1.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 1.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.6 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 4.9 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.1 | 0.2 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 4.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 1.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.9 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 1.5 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 1.0 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.3 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 1.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 1.0 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 1.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 6.2 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.9 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 2.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 2.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.8 | 3.4 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.3 | 1.0 | GO:0099569 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.2 | 1.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 1.7 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 2.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.8 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 4.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 1.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.5 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 1.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 2.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 1.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 3.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 2.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 3.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.5 | 3.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.3 | 1.7 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 2.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 0.8 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.2 | 2.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 3.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 0.8 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 1.9 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 1.0 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.6 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.5 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.4 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.5 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.8 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.9 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.3 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.7 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 1.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 2.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 1.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 3.6 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.5 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 1.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 4.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 4.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 4.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 5.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.9 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 3.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |