Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
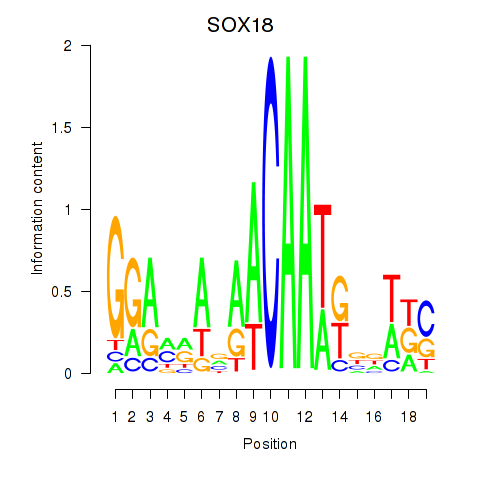
Results for SOX18
Z-value: 0.86
Transcription factors associated with SOX18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX18
|
ENSG00000203883.7 | SRY-box transcription factor 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX18 | hg38_v1_chr20_-_64049631_64049646 | -0.24 | 1.9e-01 | Click! |
Activity profile of SOX18 motif
Sorted Z-values of SOX18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_137499293 | 1.57 |
ENST00000510689.5
ENST00000394945.6 |
SPOCK1
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 1 |
| chr3_+_111998739 | 1.29 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr1_-_156248084 | 1.18 |
ENST00000652405.1
ENST00000335852.6 ENST00000540423.5 ENST00000612424.4 ENST00000613336.4 ENST00000623241.3 |
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr14_+_22070548 | 1.00 |
ENST00000390450.3
|
TRAV22
|
T cell receptor alpha variable 22 |
| chr3_+_19148500 | 1.00 |
ENST00000328405.7
|
KCNH8
|
potassium voltage-gated channel subfamily H member 8 |
| chr5_+_157180816 | 1.00 |
ENST00000422843.8
|
ITK
|
IL2 inducible T cell kinase |
| chr1_-_156248038 | 0.99 |
ENST00000470198.5
ENST00000292291.10 ENST00000356983.7 |
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr1_-_156248013 | 0.96 |
ENST00000368270.2
|
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr12_+_15322529 | 0.94 |
ENST00000348962.7
|
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr3_+_111998915 | 0.93 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr15_-_79090760 | 0.93 |
ENST00000419573.7
ENST00000558480.7 |
RASGRF1
|
Ras protein specific guanine nucleotide releasing factor 1 |
| chr5_-_147081428 | 0.90 |
ENST00000394413.7
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr12_-_10409757 | 0.88 |
ENST00000309384.2
|
KLRC4
|
killer cell lectin like receptor C4 |
| chr12_-_10435940 | 0.88 |
ENST00000381901.5
ENST00000381902.7 ENST00000539033.1 |
KLRC2
ENSG00000255641.1
|
killer cell lectin like receptor C2 novel protein |
| chr13_-_28100556 | 0.86 |
ENST00000241453.12
|
FLT3
|
fms related receptor tyrosine kinase 3 |
| chr1_+_209756032 | 0.84 |
ENST00000400959.7
ENST00000367025.8 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr4_+_69096494 | 0.82 |
ENST00000508661.5
ENST00000622664.1 |
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr3_+_186717348 | 0.82 |
ENST00000447445.1
ENST00000287611.8 ENST00000644859.2 |
KNG1
|
kininogen 1 |
| chr12_-_10420550 | 0.81 |
ENST00000381903.2
ENST00000396439.7 |
KLRC3
|
killer cell lectin like receptor C3 |
| chrM_+_8366 | 0.81 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase membrane subunit 8 |
| chr2_+_233712905 | 0.80 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr1_+_160081529 | 0.79 |
ENST00000368088.4
|
KCNJ9
|
potassium inwardly rectifying channel subfamily J member 9 |
| chr7_+_142352802 | 0.79 |
ENST00000634605.1
|
TRBV7-2
|
T cell receptor beta variable 7-2 |
| chr12_-_75209701 | 0.78 |
ENST00000350228.6
ENST00000298972.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr1_-_230745574 | 0.78 |
ENST00000681269.1
|
AGT
|
angiotensinogen |
| chr4_+_42397473 | 0.77 |
ENST00000319234.5
|
SHISA3
|
shisa family member 3 |
| chr12_-_75209814 | 0.76 |
ENST00000549446.6
|
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr14_+_21868822 | 0.75 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr1_+_209756149 | 0.75 |
ENST00000367026.7
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr6_+_24126186 | 0.72 |
ENST00000378478.5
ENST00000378491.9 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr5_-_147081462 | 0.69 |
ENST00000508267.5
ENST00000504198.5 |
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr12_+_861124 | 0.69 |
ENST00000676347.1
|
WNK1
|
WNK lysine deficient protein kinase 1 |
| chr5_-_142644201 | 0.65 |
ENST00000394496.2
ENST00000610990.4 |
FGF1
|
fibroblast growth factor 1 |
| chr17_-_40885232 | 0.64 |
ENST00000167588.4
|
KRT20
|
keratin 20 |
| chr1_-_157777124 | 0.64 |
ENST00000361516.8
ENST00000368181.4 |
FCRL2
|
Fc receptor like 2 |
| chr4_+_69096467 | 0.63 |
ENST00000305231.12
|
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr7_+_142511614 | 0.62 |
ENST00000390377.1
|
TRBV7-7
|
T cell receptor beta variable 7-7 |
| chr1_-_119811458 | 0.62 |
ENST00000256585.10
ENST00000354219.5 ENST00000369401.4 |
REG4
|
regenerating family member 4 |
| chr13_-_51845169 | 0.61 |
ENST00000627246.3
ENST00000629372.3 |
TMEM272
|
transmembrane protein 272 |
| chr12_-_110445540 | 0.60 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex subunit 3 |
| chr2_-_37672178 | 0.60 |
ENST00000457889.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr3_+_124384757 | 0.58 |
ENST00000684374.1
|
KALRN
|
kalirin RhoGEF kinase |
| chr19_+_19238259 | 0.58 |
ENST00000588231.1
|
NCAN
|
neurocan |
| chr17_-_78723556 | 0.57 |
ENST00000587308.5
|
CYTH1
|
cytohesin 1 |
| chr12_-_663572 | 0.56 |
ENST00000662884.1
|
NINJ2
|
ninjurin 2 |
| chr3_-_24495532 | 0.55 |
ENST00000643772.1
ENST00000642307.1 ENST00000645139.1 |
THRB
|
thyroid hormone receptor beta |
| chr11_-_89490715 | 0.55 |
ENST00000528341.5
|
NOX4
|
NADPH oxidase 4 |
| chr11_-_89491131 | 0.54 |
ENST00000343727.9
ENST00000531342.5 ENST00000375979.7 |
NOX4
|
NADPH oxidase 4 |
| chr2_+_79025696 | 0.54 |
ENST00000272324.10
|
REG3G
|
regenerating family member 3 gamma |
| chr4_-_173399102 | 0.53 |
ENST00000296506.8
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chr7_-_36724380 | 0.53 |
ENST00000617267.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr2_+_44275457 | 0.53 |
ENST00000611973.4
ENST00000409387.5 |
SLC3A1
|
solute carrier family 3 member 1 |
| chr11_+_73647645 | 0.53 |
ENST00000545798.5
ENST00000539157.5 ENST00000546251.5 ENST00000535582.5 ENST00000538227.5 |
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr14_+_22422371 | 0.52 |
ENST00000390469.2
|
TRDV2
|
T cell receptor delta variable 2 |
| chr13_-_51845136 | 0.52 |
ENST00000626362.3
|
TMEM272
|
transmembrane protein 272 |
| chr9_-_134917872 | 0.52 |
ENST00000616356.4
ENST00000371806.4 |
FCN1
|
ficolin 1 |
| chr11_-_105023136 | 0.52 |
ENST00000526056.5
ENST00000531367.5 ENST00000456094.1 ENST00000444749.6 ENST00000393141.6 ENST00000418434.5 ENST00000260315.8 |
CASP5
|
caspase 5 |
| chr19_-_50639827 | 0.51 |
ENST00000593901.5
ENST00000600079.6 |
SYT3
|
synaptotagmin 3 |
| chr2_+_79025678 | 0.51 |
ENST00000393897.6
|
REG3G
|
regenerating family member 3 gamma |
| chr2_-_37671633 | 0.50 |
ENST00000295324.4
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr2_+_209653171 | 0.50 |
ENST00000447185.5
|
MAP2
|
microtubule associated protein 2 |
| chr3_+_68006224 | 0.50 |
ENST00000496687.1
|
TAFA1
|
TAFA chemokine like family member 1 |
| chr7_+_142529268 | 0.50 |
ENST00000612787.1
|
TRBV7-9
|
T cell receptor beta variable 7-9 |
| chr1_-_182391783 | 0.49 |
ENST00000331872.11
ENST00000339526.8 |
GLUL
|
glutamate-ammonia ligase |
| chr2_+_79025709 | 0.48 |
ENST00000409471.1
|
REG3G
|
regenerating family member 3 gamma |
| chr5_-_67196791 | 0.47 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr1_+_179954740 | 0.47 |
ENST00000491495.2
ENST00000367607.8 |
CEP350
|
centrosomal protein 350 |
| chr11_+_73648889 | 0.47 |
ENST00000542389.5
|
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr11_+_13277639 | 0.46 |
ENST00000527998.5
ENST00000529388.6 ENST00000401424.6 ENST00000403510.8 ENST00000403290.6 ENST00000533520.5 ENST00000389707.8 ENST00000673817.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator like |
| chr7_-_123199960 | 0.46 |
ENST00000194130.7
|
SLC13A1
|
solute carrier family 13 member 1 |
| chr3_+_124384772 | 0.46 |
ENST00000682575.1
|
KALRN
|
kalirin RhoGEF kinase |
| chr12_-_95996302 | 0.45 |
ENST00000261208.8
ENST00000538703.5 ENST00000541929.5 |
HAL
|
histidine ammonia-lyase |
| chr12_-_262828 | 0.45 |
ENST00000343164.9
ENST00000436453.1 ENST00000445055.6 ENST00000546319.5 |
SLC6A13
|
solute carrier family 6 member 13 |
| chr12_-_75209422 | 0.44 |
ENST00000393288.2
ENST00000540018.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chrX_-_149505274 | 0.44 |
ENST00000428056.6
ENST00000340855.11 ENST00000370441.8 |
IDS
|
iduronate 2-sulfatase |
| chr1_-_182391363 | 0.44 |
ENST00000417584.6
|
GLUL
|
glutamate-ammonia ligase |
| chr10_+_88759997 | 0.43 |
ENST00000404459.2
|
LIPN
|
lipase family member N |
| chr17_-_75855204 | 0.42 |
ENST00000589642.5
ENST00000593002.1 ENST00000590221.5 ENST00000587374.5 ENST00000585462.5 ENST00000254806.8 ENST00000433525.6 ENST00000626827.2 |
WBP2
|
WW domain binding protein 2 |
| chr2_+_44275491 | 0.42 |
ENST00000410056.7
ENST00000409741.5 ENST00000409229.7 |
SLC3A1
|
solute carrier family 3 member 1 |
| chr12_-_110689751 | 0.42 |
ENST00000439744.6
ENST00000549442.5 |
HVCN1
|
hydrogen voltage gated channel 1 |
| chr1_+_240091866 | 0.41 |
ENST00000319653.14
|
FMN2
|
formin 2 |
| chr17_-_17281232 | 0.41 |
ENST00000417352.5
ENST00000268717.10 |
COPS3
|
COP9 signalosome subunit 3 |
| chr19_+_4402685 | 0.41 |
ENST00000585854.1
|
CHAF1A
|
chromatin assembly factor 1 subunit A |
| chr13_-_46142834 | 0.40 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr14_+_22096017 | 0.40 |
ENST00000390452.2
|
TRDV1
|
T cell receptor delta variable 1 |
| chrX_+_86714623 | 0.40 |
ENST00000484479.1
|
DACH2
|
dachshund family transcription factor 2 |
| chrX_-_154805386 | 0.40 |
ENST00000393531.5
ENST00000369534.8 ENST00000453245.5 ENST00000428488.1 ENST00000369531.1 |
MPP1
|
membrane palmitoylated protein 1 |
| chr9_+_33240159 | 0.40 |
ENST00000379721.4
|
SPINK4
|
serine peptidase inhibitor Kazal type 4 |
| chr17_-_45425620 | 0.39 |
ENST00000376922.6
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr14_+_21768482 | 0.39 |
ENST00000390428.3
|
TRAV6
|
T cell receptor alpha variable 6 |
| chr12_+_64824602 | 0.39 |
ENST00000539867.6
ENST00000544457.1 ENST00000539120.1 |
TBC1D30
|
TBC1 domain family member 30 |
| chr1_+_99850348 | 0.39 |
ENST00000361915.8
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr1_+_247605586 | 0.39 |
ENST00000320002.3
|
OR2G3
|
olfactory receptor family 2 subfamily G member 3 |
| chr4_+_37960397 | 0.39 |
ENST00000504686.2
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr13_-_83882390 | 0.39 |
ENST00000377084.3
|
SLITRK1
|
SLIT and NTRK like family member 1 |
| chr13_+_46212002 | 0.39 |
ENST00000676446.1
|
LRRC63
|
leucine rich repeat containing 63 |
| chr19_-_50639282 | 0.39 |
ENST00000598997.1
|
SYT3
|
synaptotagmin 3 |
| chr11_+_73648979 | 0.38 |
ENST00000540431.1
|
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr6_+_42915989 | 0.38 |
ENST00000441198.4
ENST00000446507.5 ENST00000616441.2 |
PTCRA
|
pre T cell antigen receptor alpha |
| chr1_-_156859087 | 0.38 |
ENST00000368195.4
|
INSRR
|
insulin receptor related receptor |
| chr18_+_35041387 | 0.38 |
ENST00000538170.6
ENST00000300249.10 ENST00000588910.5 |
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr7_-_93574721 | 0.38 |
ENST00000426151.7
ENST00000649521.1 |
CALCR
|
calcitonin receptor |
| chr3_-_121660892 | 0.37 |
ENST00000428394.6
ENST00000314583.8 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr16_-_68000549 | 0.37 |
ENST00000575510.5
|
DPEP2
|
dipeptidase 2 |
| chr8_-_101206064 | 0.37 |
ENST00000518336.5
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr3_+_160677152 | 0.37 |
ENST00000320767.4
|
ARL14
|
ADP ribosylation factor like GTPase 14 |
| chr19_-_51501755 | 0.37 |
ENST00000291707.8
|
SIGLEC12
|
sialic acid binding Ig like lectin 12 |
| chr1_-_150765735 | 0.37 |
ENST00000679898.1
ENST00000448301.7 ENST00000680664.1 ENST00000679512.1 ENST00000368985.8 ENST00000679582.1 |
CTSS
|
cathepsin S |
| chr3_+_124384513 | 0.36 |
ENST00000682540.1
ENST00000522553.6 ENST00000682695.1 ENST00000682674.1 ENST00000684382.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr5_-_74640649 | 0.36 |
ENST00000537006.1
|
ENC1
|
ectodermal-neural cortex 1 |
| chr9_-_71911183 | 0.35 |
ENST00000333421.7
|
ABHD17B
|
abhydrolase domain containing 17B, depalmitoylase |
| chr2_+_240605417 | 0.35 |
ENST00000319838.10
ENST00000403859.1 |
GPR35
|
G protein-coupled receptor 35 |
| chr1_+_54806063 | 0.35 |
ENST00000358193.7
ENST00000371273.4 |
LEXM
|
lymphocyte expansion molecule |
| chr12_-_55927756 | 0.35 |
ENST00000549939.1
|
PYM1
|
PYM homolog 1, exon junction complex associated factor |
| chr5_-_59356962 | 0.35 |
ENST00000405755.6
|
PDE4D
|
phosphodiesterase 4D |
| chr9_-_127735286 | 0.34 |
ENST00000336067.10
ENST00000373284.10 ENST00000373281.8 ENST00000463577.2 |
TOR2A
|
torsin family 2 member A |
| chr5_+_31532277 | 0.34 |
ENST00000507818.6
ENST00000325366.14 |
C5orf22
|
chromosome 5 open reading frame 22 |
| chr9_+_135702172 | 0.34 |
ENST00000487664.5
ENST00000371757.7 ENST00000628528.2 |
KCNT1
|
potassium sodium-activated channel subfamily T member 1 |
| chr12_-_70754631 | 0.34 |
ENST00000440835.6
ENST00000549308.5 ENST00000550661.1 ENST00000378778.5 |
PTPRR
|
protein tyrosine phosphatase receptor type R |
| chr6_+_25652272 | 0.34 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr16_-_15643024 | 0.34 |
ENST00000540441.6
|
MARF1
|
meiosis regulator and mRNA stability factor 1 |
| chr15_+_62066975 | 0.33 |
ENST00000355522.5
|
C2CD4A
|
C2 calcium dependent domain containing 4A |
| chr12_+_110502307 | 0.33 |
ENST00000409778.7
ENST00000409300.6 |
RAD9B
|
RAD9 checkpoint clamp component B |
| chr9_+_135095978 | 0.33 |
ENST00000545657.1
|
OLFM1
|
olfactomedin 1 |
| chr8_+_85987287 | 0.33 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase H+ transporting V0 subunit d2 |
| chr12_+_69239627 | 0.33 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6 |
| chrM_+_8489 | 0.33 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase membrane subunit 6 |
| chr22_+_44026283 | 0.32 |
ENST00000619710.4
|
PARVB
|
parvin beta |
| chr15_+_76995374 | 0.32 |
ENST00000559161.5
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr18_-_5396265 | 0.32 |
ENST00000579951.2
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr9_-_21351378 | 0.31 |
ENST00000380210.1
|
IFNA6
|
interferon alpha 6 |
| chr2_-_36966503 | 0.31 |
ENST00000263918.9
|
STRN
|
striatin |
| chr16_+_20451532 | 0.31 |
ENST00000576361.5
|
ACSM2A
|
acyl-CoA synthetase medium chain family member 2A |
| chr2_-_106160106 | 0.30 |
ENST00000479774.5
|
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chr2_+_100974849 | 0.30 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr7_+_6009222 | 0.30 |
ENST00000400479.6
ENST00000223029.8 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex interacting multifunctional protein 2 |
| chr11_-_31804067 | 0.30 |
ENST00000639548.1
ENST00000640125.1 ENST00000481563.6 ENST00000639079.1 ENST00000638762.1 ENST00000638346.1 |
PAX6
|
paired box 6 |
| chr19_+_54630497 | 0.30 |
ENST00000396332.8
ENST00000427581.6 |
LILRB1
|
leukocyte immunoglobulin like receptor B1 |
| chr15_-_80252205 | 0.30 |
ENST00000560778.3
|
CTXND1
|
cortexin domain containing 1 |
| chr1_-_72100930 | 0.30 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr12_-_7751605 | 0.29 |
ENST00000354629.9
|
CLEC4C
|
C-type lectin domain family 4 member C |
| chr6_-_111793900 | 0.29 |
ENST00000462598.7
|
FYN
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr11_+_65181920 | 0.29 |
ENST00000279247.11
ENST00000532285.1 ENST00000534373.5 |
CAPN1
|
calpain 1 |
| chr1_+_99850485 | 0.29 |
ENST00000370165.7
ENST00000370163.7 ENST00000294724.8 |
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr5_-_79512794 | 0.29 |
ENST00000282260.10
ENST00000508576.5 ENST00000535690.1 |
HOMER1
|
homer scaffold protein 1 |
| chr1_-_209784521 | 0.29 |
ENST00000294811.2
|
C1orf74
|
chromosome 1 open reading frame 74 |
| chr5_+_149581368 | 0.28 |
ENST00000333677.7
|
ARHGEF37
|
Rho guanine nucleotide exchange factor 37 |
| chr5_-_56116946 | 0.28 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr16_-_90019414 | 0.28 |
ENST00000002501.11
|
DBNDD1
|
dysbindin domain containing 1 |
| chr12_-_69699331 | 0.27 |
ENST00000548658.1
ENST00000476098.5 |
BEST3
|
bestrophin 3 |
| chr13_-_52450590 | 0.27 |
ENST00000378060.9
|
VPS36
|
vacuolar protein sorting 36 homolog |
| chr12_-_69699382 | 0.27 |
ENST00000551160.5
|
BEST3
|
bestrophin 3 |
| chr12_+_69239592 | 0.27 |
ENST00000456847.7
ENST00000266679.8 |
CPSF6
|
cleavage and polyadenylation specific factor 6 |
| chr3_-_112974912 | 0.27 |
ENST00000440122.6
ENST00000490004.1 |
CD200R1
|
CD200 receptor 1 |
| chr2_+_44275473 | 0.27 |
ENST00000260649.11
|
SLC3A1
|
solute carrier family 3 member 1 |
| chr12_-_69699353 | 0.27 |
ENST00000331471.8
|
BEST3
|
bestrophin 3 |
| chr20_+_841238 | 0.27 |
ENST00000541082.2
|
FAM110A
|
family with sequence similarity 110 member A |
| chr17_-_30334050 | 0.27 |
ENST00000328886.5
ENST00000538566.6 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr11_-_59713845 | 0.26 |
ENST00000307552.3
|
OR10V1
|
olfactory receptor family 10 subfamily V member 1 |
| chr4_-_122456725 | 0.26 |
ENST00000226730.5
|
IL2
|
interleukin 2 |
| chr6_+_42916046 | 0.26 |
ENST00000304672.6
|
PTCRA
|
pre T cell antigen receptor alpha |
| chr22_+_37282464 | 0.26 |
ENST00000402997.5
ENST00000405206.3 ENST00000248901.11 |
CYTH4
|
cytohesin 4 |
| chrX_-_111412162 | 0.26 |
ENST00000637570.1
ENST00000356220.8 ENST00000636035.2 ENST00000637453.1 ENST00000635795.1 |
DCX
|
doublecortin |
| chr22_-_38084093 | 0.26 |
ENST00000681075.1
|
SLC16A8
|
solute carrier family 16 member 8 |
| chr3_+_124384376 | 0.25 |
ENST00000682636.1
|
KALRN
|
kalirin RhoGEF kinase |
| chr19_+_7049321 | 0.25 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3 like 2 |
| chrX_+_3066833 | 0.24 |
ENST00000359361.2
|
ARSF
|
arylsulfatase F |
| chr19_-_41688258 | 0.24 |
ENST00000401731.6
ENST00000006724.7 |
CEACAM7
|
CEA cell adhesion molecule 7 |
| chr7_+_157336988 | 0.23 |
ENST00000262177.9
ENST00000417758.5 ENST00000443280.5 |
DNAJB6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr12_-_69699285 | 0.23 |
ENST00000553096.5
ENST00000330891.10 |
BEST3
|
bestrophin 3 |
| chr11_+_65182413 | 0.23 |
ENST00000531068.5
ENST00000527699.5 ENST00000533909.5 ENST00000527323.5 |
CAPN1
|
calpain 1 |
| chr12_-_11092313 | 0.23 |
ENST00000531678.1
|
TAS2R43
|
taste 2 receptor member 43 |
| chrX_-_47145035 | 0.22 |
ENST00000276062.8
|
NDUFB11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr13_+_111115303 | 0.22 |
ENST00000646102.2
ENST00000449979.5 |
ARHGEF7
|
Rho guanine nucleotide exchange factor 7 |
| chr11_+_5689691 | 0.22 |
ENST00000425490.5
|
TRIM22
|
tripartite motif containing 22 |
| chr15_-_68205319 | 0.22 |
ENST00000467889.3
ENST00000448060.7 |
CALML4
|
calmodulin like 4 |
| chr11_+_74988896 | 0.22 |
ENST00000526068.1
ENST00000294064.9 ENST00000532963.1 ENST00000531619.1 ENST00000534628.1 |
NEU3
|
neuraminidase 3 |
| chr8_+_24294044 | 0.22 |
ENST00000265769.9
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr15_-_55249029 | 0.22 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_-_11858935 | 0.22 |
ENST00000376468.4
|
NPPB
|
natriuretic peptide B |
| chr2_-_240821363 | 0.22 |
ENST00000675940.1
|
KIF1A
|
kinesin family member 1A |
| chr14_+_21749163 | 0.22 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr1_-_155911365 | 0.22 |
ENST00000651833.1
ENST00000539040.5 ENST00000651853.1 |
RIT1
|
Ras like without CAAX 1 |
| chr6_-_26234978 | 0.22 |
ENST00000244534.7
|
H1-3
|
H1.3 linker histone, cluster member |
| chr7_-_33040893 | 0.21 |
ENST00000643244.1
ENST00000409467.6 ENST00000449201.5 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr13_+_31739542 | 0.21 |
ENST00000380314.2
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr19_+_8052752 | 0.21 |
ENST00000315626.6
ENST00000253451.9 |
CCL25
|
C-C motif chemokine ligand 25 |
| chr4_+_153466324 | 0.21 |
ENST00000409663.7
ENST00000409959.8 |
TMEM131L
|
transmembrane 131 like |
| chr16_+_24539536 | 0.21 |
ENST00000568015.5
ENST00000319715.10 |
RBBP6
|
RB binding protein 6, ubiquitin ligase |
| chr1_-_248645278 | 0.21 |
ENST00000641268.1
|
OR2T35
|
olfactory receptor family 2 subfamily T member 35 |
| chr19_-_45602153 | 0.21 |
ENST00000544371.1
ENST00000323040.5 |
OPA3
GPR4
|
outer mitochondrial membrane lipid metabolism regulator OPA3 G protein-coupled receptor 4 |
| chr3_-_47891269 | 0.21 |
ENST00000335271.9
|
MAP4
|
microtubule associated protein 4 |
| chr16_-_90019821 | 0.20 |
ENST00000568838.2
|
DBNDD1
|
dysbindin domain containing 1 |
| chr12_-_8066331 | 0.20 |
ENST00000546241.1
ENST00000307637.5 |
C3AR1
|
complement C3a receptor 1 |
| chr6_+_155013646 | 0.20 |
ENST00000538270.5
ENST00000535231.5 |
TIAM2
|
TIAM Rac1 associated GEF 2 |
| chr17_+_58238694 | 0.20 |
ENST00000581008.1
|
LPO
|
lactoperoxidase |
| chr2_+_74654304 | 0.20 |
ENST00000453930.5
|
SEMA4F
|
ssemaphorin 4F |
| chrX_-_47144680 | 0.19 |
ENST00000377811.4
|
NDUFB11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr8_+_24294107 | 0.19 |
ENST00000437154.6
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr12_-_110502065 | 0.19 |
ENST00000447578.6
ENST00000546588.1 ENST00000360579.11 ENST00000549578.6 ENST00000549970.5 |
VPS29
|
VPS29 retromer complex component |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX18
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.5 | 2.0 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.3 | 0.8 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.2 | 0.9 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 0.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 1.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.2 | 0.5 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 1.7 | GO:0060125 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 1.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.4 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.4 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.3 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.1 | 0.7 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.4 | GO:0002290 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.1 | 0.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 2.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.5 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 1.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.4 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.3 | GO:0033320 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.1 | 0.5 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.6 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.5 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.6 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 | 1.0 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.3 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.5 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 1.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.6 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 1.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.4 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.4 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.7 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.4 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.2 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.9 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.2 | GO:0072143 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.4 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.3 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.3 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.3 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 1.0 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.5 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 1.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.5 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.3 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.5 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.7 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 2.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.4 | GO:0045651 | positive regulation of granulocyte differentiation(GO:0030854) positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.4 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.2 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.0 | 0.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.8 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.1 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.6 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) coenzyme transport(GO:0051182) |
| 0.0 | 0.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.5 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.5 | GO:0097169 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 2.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 1.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.9 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.3 | GO:0033177 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 1.2 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 0.9 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 0.5 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.2 | 0.9 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 1.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.4 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.1 | 1.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 1.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.4 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.3 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.1 | 1.5 | GO:0070492 | peptidoglycan binding(GO:0042834) oligosaccharide binding(GO:0070492) |
| 0.1 | 0.5 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.4 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 2.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 1.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.7 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 2.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 1.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.5 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 2.9 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 2.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 3.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |