Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
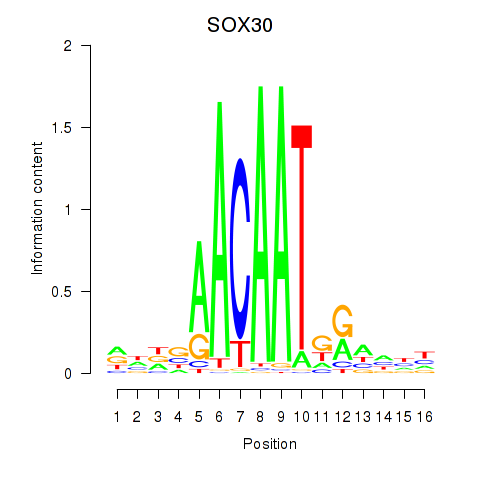
Results for SOX30
Z-value: 1.05
Transcription factors associated with SOX30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX30
|
ENSG00000039600.11 | SRY-box transcription factor 30 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX30 | hg38_v1_chr5_-_157652364_157652385 | 0.03 | 8.8e-01 | Click! |
Activity profile of SOX30 motif
Sorted Z-values of SOX30 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_127900958 | 3.59 |
ENST00000434358.3
ENST00000630369.2 ENST00000368248.4 |
THEMIS
|
thymocyte selection associated |
| chr11_-_35419542 | 3.36 |
ENST00000643305.1
ENST00000644351.1 ENST00000278379.9 ENST00000644779.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr4_-_175812746 | 3.34 |
ENST00000393658.6
|
GPM6A
|
glycoprotein M6A |
| chr2_-_157325808 | 3.31 |
ENST00000410096.6
ENST00000420719.6 ENST00000409216.5 ENST00000419116.2 |
ERMN
|
ermin |
| chr11_-_35420050 | 2.79 |
ENST00000395753.6
ENST00000395750.6 ENST00000645634.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr8_+_79611036 | 2.78 |
ENST00000220876.12
ENST00000518111.5 |
STMN2
|
stathmin 2 |
| chr5_-_147081428 | 2.70 |
ENST00000394413.7
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr11_-_35419213 | 2.62 |
ENST00000642171.1
ENST00000644050.1 ENST00000643134.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr12_-_98894830 | 2.61 |
ENST00000549797.5
ENST00000333732.11 ENST00000341752.11 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr3_+_181711915 | 2.53 |
ENST00000325404.3
|
SOX2
|
SRY-box transcription factor 2 |
| chr11_-_35419462 | 2.34 |
ENST00000643522.1
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr5_-_147081462 | 2.34 |
ENST00000508267.5
ENST00000504198.5 |
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr11_-_35420017 | 2.26 |
ENST00000643000.1
ENST00000646099.1 ENST00000647372.1 ENST00000642578.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr12_-_11395556 | 2.20 |
ENST00000565533.1
ENST00000389362.6 ENST00000546254.3 |
PRB2
PRB1
|
proline rich protein BstNI subfamily 2 proline rich protein BstNI subfamily 1 |
| chr1_-_25905470 | 2.12 |
ENST00000446334.1
|
STMN1
|
stathmin 1 |
| chr11_-_35419899 | 2.06 |
ENST00000646847.1
ENST00000449068.2 ENST00000643401.1 ENST00000645966.1 ENST00000647104.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr12_-_11310420 | 1.95 |
ENST00000621732.4
ENST00000445719.2 ENST00000279575.7 |
PRB4
|
proline rich protein BstNI subfamily 4 |
| chr17_-_40665121 | 1.89 |
ENST00000394052.5
|
KRT222
|
keratin 222 |
| chr2_-_240820945 | 1.88 |
ENST00000428768.2
ENST00000650053.1 ENST00000650130.1 |
KIF1A
|
kinesin family member 1A |
| chr11_-_85719313 | 1.87 |
ENST00000526999.5
|
SYTL2
|
synaptotagmin like 2 |
| chr9_-_131270493 | 1.82 |
ENST00000372269.7
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78 member A |
| chr15_+_92883413 | 1.82 |
ENST00000629685.2
|
ENSG00000279765.3
|
novel protein |
| chr21_+_29300111 | 1.78 |
ENST00000451655.5
|
BACH1
|
BTB domain and CNC homolog 1 |
| chr12_-_49187369 | 1.77 |
ENST00000547939.6
|
TUBA1A
|
tubulin alpha 1a |
| chrX_-_24672654 | 1.76 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr8_-_81112055 | 1.70 |
ENST00000220597.4
|
PAG1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr7_+_142352802 | 1.62 |
ENST00000634605.1
|
TRBV7-2
|
T cell receptor beta variable 7-2 |
| chr1_-_25905098 | 1.59 |
ENST00000374291.5
|
STMN1
|
stathmin 1 |
| chr12_+_10929229 | 1.55 |
ENST00000381847.7
ENST00000396400.4 |
PRH2
|
proline rich protein HaeIII subfamily 2 |
| chr12_-_10130241 | 1.52 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr12_-_98895263 | 1.51 |
ENST00000548447.1
ENST00000546364.7 ENST00000552748.5 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr17_-_75153826 | 1.46 |
ENST00000481647.5
ENST00000470924.5 |
JPT1
|
Jupiter microtubule associated homolog 1 |
| chrX_-_143636094 | 1.45 |
ENST00000356928.2
|
SLITRK4
|
SLIT and NTRK like family member 4 |
| chr1_+_154429315 | 1.43 |
ENST00000476006.5
|
IL6R
|
interleukin 6 receptor |
| chr12_+_50504970 | 1.42 |
ENST00000301180.10
|
DIP2B
|
disco interacting protein 2 homolog B |
| chr21_+_29299368 | 1.40 |
ENST00000399921.5
|
BACH1
|
BTB domain and CNC homolog 1 |
| chr1_-_243255320 | 1.38 |
ENST00000366544.5
ENST00000366543.5 |
CEP170
|
centrosomal protein 170 |
| chr12_-_98895199 | 1.35 |
ENST00000552407.5
ENST00000551613.5 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr22_+_50170720 | 1.34 |
ENST00000159647.9
ENST00000395842.3 |
PANX2
|
pannexin 2 |
| chrX_-_15854743 | 1.29 |
ENST00000450644.2
|
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chrX_-_15854791 | 1.26 |
ENST00000545766.7
ENST00000380291.5 ENST00000672987.1 ENST00000329235.6 |
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chr1_-_243255170 | 1.22 |
ENST00000366542.6
|
CEP170
|
centrosomal protein 170 |
| chr8_+_42338454 | 1.19 |
ENST00000532157.5
ENST00000520008.5 |
POLB
|
DNA polymerase beta |
| chr12_-_11269696 | 1.18 |
ENST00000381842.7
|
PRB3
|
proline rich protein BstNI subfamily 3 |
| chr12_-_11269805 | 1.16 |
ENST00000538488.2
|
PRB3
|
proline rich protein BstNI subfamily 3 |
| chr10_-_60300474 | 1.15 |
ENST00000503925.1
|
ANK3
|
ankyrin 3 |
| chr3_+_147393718 | 1.15 |
ENST00000488404.5
|
ZIC1
|
Zic family member 1 |
| chr11_-_85719045 | 1.13 |
ENST00000533057.6
ENST00000533892.5 |
SYTL2
|
synaptotagmin like 2 |
| chr12_-_10130143 | 1.13 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr7_-_14903319 | 1.10 |
ENST00000403951.6
|
DGKB
|
diacylglycerol kinase beta |
| chr7_+_142670734 | 1.10 |
ENST00000390398.3
|
TRBV25-1
|
T cell receptor beta variable 25-1 |
| chr9_+_100473140 | 1.02 |
ENST00000374879.5
|
TMEFF1
|
transmembrane protein with EGF like and two follistatin like domains 1 |
| chr1_+_240091866 | 1.02 |
ENST00000319653.14
|
FMN2
|
formin 2 |
| chr11_-_85719160 | 1.01 |
ENST00000389958.7
ENST00000527794.5 |
SYTL2
|
synaptotagmin like 2 |
| chr11_-_85719111 | 1.01 |
ENST00000529581.5
ENST00000533577.1 |
SYTL2
|
synaptotagmin like 2 |
| chr17_+_69502154 | 0.94 |
ENST00000589295.5
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr3_-_18424533 | 0.90 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr15_-_48963912 | 0.89 |
ENST00000332408.9
|
SHC4
|
SHC adaptor protein 4 |
| chr17_+_69502397 | 0.86 |
ENST00000613873.4
ENST00000589647.5 |
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr2_-_165203870 | 0.86 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr3_-_13042419 | 0.86 |
ENST00000647458.1
|
IQSEC1
|
IQ motif and Sec7 domain ArfGEF 1 |
| chr11_+_36296281 | 0.84 |
ENST00000530639.6
|
PRR5L
|
proline rich 5 like |
| chr1_-_204494701 | 0.83 |
ENST00000429009.1
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr11_+_128693887 | 0.82 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr8_+_42338477 | 0.81 |
ENST00000518925.5
ENST00000265421.9 |
POLB
|
DNA polymerase beta |
| chr11_+_128694052 | 0.79 |
ENST00000527786.7
ENST00000534087.3 |
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_-_83673544 | 0.77 |
ENST00000503117.1
ENST00000510978.5 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr12_-_10130082 | 0.76 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A |
| chr4_+_143381939 | 0.72 |
ENST00000505913.5
|
GAB1
|
GRB2 associated binding protein 1 |
| chr17_+_69502255 | 0.71 |
ENST00000588110.5
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_+_74235377 | 0.66 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr1_-_243255348 | 0.65 |
ENST00000522995.1
|
CEP170
|
centrosomal protein 170 |
| chr2_-_165204042 | 0.64 |
ENST00000283254.12
ENST00000453007.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr10_-_27981805 | 0.64 |
ENST00000673512.1
ENST00000672877.1 ENST00000480504.1 |
ODAD2
|
outer dynein arm docking complex subunit 2 |
| chr5_+_68292486 | 0.64 |
ENST00000519025.5
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr2_-_37672178 | 0.64 |
ENST00000457889.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr2_+_178320228 | 0.63 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr7_-_83162899 | 0.61 |
ENST00000423517.6
|
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr6_+_125919296 | 0.59 |
ENST00000444128.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr5_-_160852200 | 0.58 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr5_-_135399211 | 0.53 |
ENST00000312469.8
ENST00000423969.6 |
MACROH2A1
|
macroH2A.1 histone |
| chr11_-_102452758 | 0.53 |
ENST00000398136.7
ENST00000361236.7 |
TMEM123
|
transmembrane protein 123 |
| chr15_+_76336755 | 0.51 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr3_+_141262614 | 0.51 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chr2_-_240821363 | 0.49 |
ENST00000675940.1
|
KIF1A
|
kinesin family member 1A |
| chr2_-_37671633 | 0.49 |
ENST00000295324.4
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr6_+_125919210 | 0.49 |
ENST00000438495.6
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr2_+_86441995 | 0.48 |
ENST00000427678.1
|
KDM3A
|
lysine demethylase 3A |
| chr2_-_37672448 | 0.47 |
ENST00000611976.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr20_-_63979629 | 0.43 |
ENST00000369886.8
ENST00000450107.1 |
SAMD10
|
sterile alpha motif domain containing 10 |
| chr6_-_11778781 | 0.42 |
ENST00000414691.8
ENST00000229583.9 |
ADTRP
|
androgen dependent TFPI regulating protein |
| chr8_+_24294044 | 0.42 |
ENST00000265769.9
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr20_+_38962299 | 0.38 |
ENST00000373325.6
ENST00000373323.8 ENST00000252011.8 ENST00000615559.1 |
DHX35
ENSG00000277581.1
|
DEAH-box helicase 35 novel transcript, sense intronic to DHX35 |
| chr9_-_71768386 | 0.36 |
ENST00000377066.9
ENST00000377044.9 |
CEMIP2
|
cell migration inducing hyaluronidase 2 |
| chr16_+_4495852 | 0.36 |
ENST00000575129.5
ENST00000398595.7 ENST00000414777.5 |
HMOX2
|
heme oxygenase 2 |
| chr16_+_68539213 | 0.33 |
ENST00000570495.5
ENST00000564323.5 ENST00000562156.5 ENST00000573685.5 ENST00000563169.7 ENST00000611381.4 |
ZFP90
|
ZFP90 zinc finger protein |
| chr10_-_59363176 | 0.32 |
ENST00000512919.5
|
FAM13C
|
family with sequence similarity 13 member C |
| chr17_-_64390592 | 0.31 |
ENST00000563523.5
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1 |
| chr12_+_92702843 | 0.28 |
ENST00000397833.3
|
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr8_+_24294107 | 0.26 |
ENST00000437154.6
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr13_-_85799400 | 0.25 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr12_+_27470632 | 0.22 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr10_-_122699768 | 0.22 |
ENST00000432000.5
|
C10orf120
|
chromosome 10 open reading frame 120 |
| chr5_-_135399863 | 0.21 |
ENST00000510038.1
ENST00000304332.8 |
MACROH2A1
|
macroH2A.1 histone |
| chr11_+_63369779 | 0.21 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chr16_-_2787550 | 0.19 |
ENST00000682474.1
|
PRSS33
|
serine protease 33 |
| chr12_+_92702983 | 0.19 |
ENST00000344636.6
ENST00000544406.2 |
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr11_+_196761 | 0.18 |
ENST00000325113.9
|
ODF3
|
outer dense fiber of sperm tails 3 |
| chr15_+_56918763 | 0.18 |
ENST00000557843.5
|
TCF12
|
transcription factor 12 |
| chr11_+_65890627 | 0.17 |
ENST00000312579.4
|
CCDC85B
|
coiled-coil domain containing 85B |
| chr15_+_56919120 | 0.17 |
ENST00000557947.5
|
TCF12
|
transcription factor 12 |
| chr17_+_7420315 | 0.16 |
ENST00000323675.4
|
SPEM1
|
spermatid maturation 1 |
| chr10_-_122699836 | 0.15 |
ENST00000329446.5
|
C10orf120
|
chromosome 10 open reading frame 120 |
| chr12_-_95217373 | 0.14 |
ENST00000549499.1
ENST00000546711.5 ENST00000343958.9 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr6_+_12290353 | 0.14 |
ENST00000379375.6
|
EDN1
|
endothelin 1 |
| chr3_-_149221811 | 0.14 |
ENST00000455472.3
ENST00000264613.11 |
CP
|
ceruloplasmin |
| chr1_+_40477248 | 0.13 |
ENST00000372706.6
|
ZFP69
|
ZFP69 zinc finger protein |
| chr1_+_192316992 | 0.13 |
ENST00000417209.2
|
RGS21
|
regulator of G protein signaling 21 |
| chr13_-_41019289 | 0.11 |
ENST00000239882.7
|
ELF1
|
E74 like ETS transcription factor 1 |
| chrX_+_80420466 | 0.10 |
ENST00000308293.5
|
TENT5D
|
terminal nucleotidyltransferase 5D |
| chr6_-_89217339 | 0.10 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1 |
| chr8_+_7836433 | 0.09 |
ENST00000314265.3
|
DEFB104A
|
defensin beta 104A |
| chr10_-_114632011 | 0.09 |
ENST00000651023.1
|
ABLIM1
|
actin binding LIM protein 1 |
| chr1_-_167914089 | 0.08 |
ENST00000476818.2
ENST00000367848.1 ENST00000367851.9 ENST00000545172.5 |
ADCY10
|
adenylate cyclase 10 |
| chrX_+_44844015 | 0.08 |
ENST00000339042.6
|
DUSP21
|
dual specificity phosphatase 21 |
| chrX_+_102937272 | 0.07 |
ENST00000218249.7
|
RAB40AL
|
RAB40A like |
| chr3_+_19947074 | 0.07 |
ENST00000273047.9
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr1_+_65419 | 0.07 |
ENST00000641515.2
|
OR4F5
|
olfactory receptor family 4 subfamily F member 5 |
| chr19_+_107104 | 0.04 |
ENST00000585993.3
ENST00000618231.3 |
OR4F17
|
olfactory receptor family 4 subfamily F member 17 |
| chr2_+_29113989 | 0.03 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr12_+_55492378 | 0.03 |
ENST00000548615.1
|
OR6C68
|
olfactory receptor family 6 subfamily C member 68 |
| chr17_-_676348 | 0.02 |
ENST00000681510.1
ENST00000679680.1 |
VPS53
|
VPS53 subunit of GARP complex |
| chr14_+_35826298 | 0.01 |
ENST00000216807.12
|
BRMS1L
|
BRMS1 like transcriptional repressor |
| chr3_-_71306012 | 0.01 |
ENST00000649431.1
ENST00000610810.5 |
FOXP1
|
forkhead box P1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX30
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 15.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.7 | 2.8 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.7 | 5.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.5 | 3.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.5 | 2.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.4 | 2.0 | GO:0006288 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) base-excision repair, DNA ligation(GO:0006288) |
| 0.3 | 5.0 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.3 | 1.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.3 | 3.7 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.2 | 2.5 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.2 | 1.0 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.2 | 0.6 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.2 | 5.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 0.5 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.2 | 1.8 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 1.2 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 3.6 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.1 | 3.4 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.5 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 1.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 1.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 3.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 2.4 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 1.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.9 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 2.5 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0060584 | nitric oxide transport(GO:0030185) rhythmic synaptic transmission(GO:0060024) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.5 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.7 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 1.1 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 1.5 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.6 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.1 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.0 | 0.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 1.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 1.7 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 1.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 1.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 15.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 0.6 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 4.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.6 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 5.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 3.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 1.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 2.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.5 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 2.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 3.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 4.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.9 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 15.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 1.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 1.4 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.4 | 1.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 1.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 5.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 3.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 5.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.4 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 2.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 2.0 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 2.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 2.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 5.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 2.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 3.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 2.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.7 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 5.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 3.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 15.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 1.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |