Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
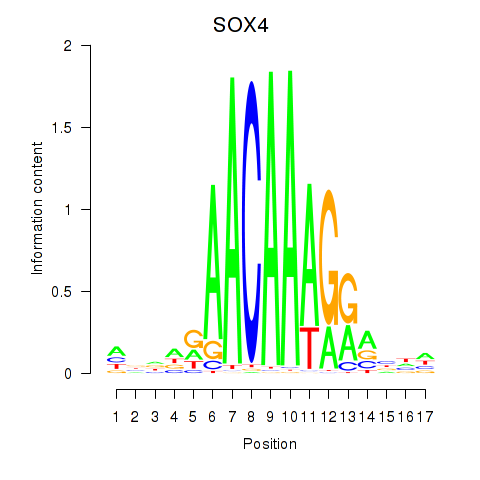
Results for SOX4
Z-value: 0.82
Transcription factors associated with SOX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX4
|
ENSG00000124766.7 | SRY-box transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX4 | hg38_v1_chr6_+_21593742_21593757 | 0.15 | 4.0e-01 | Click! |
Activity profile of SOX4 motif
Sorted Z-values of SOX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_110307131 | 2.72 |
ENST00000543140.6
ENST00000375820.10 |
COL4A1
|
collagen type IV alpha 1 chain |
| chr17_-_64390852 | 2.38 |
ENST00000563924.6
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1 |
| chr17_-_64390592 | 2.11 |
ENST00000563523.5
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1 |
| chr6_-_139291987 | 1.91 |
ENST00000358430.8
|
TXLNB
|
taxilin beta |
| chr9_+_98943705 | 1.83 |
ENST00000610452.1
|
COL15A1
|
collagen type XV alpha 1 chain |
| chr7_-_27180230 | 1.78 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr10_+_6202866 | 1.71 |
ENST00000317350.8
ENST00000379785.5 ENST00000625260.2 ENST00000626882.2 ENST00000360521.7 ENST00000379775.9 ENST00000640683.1 |
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr7_+_80638510 | 1.68 |
ENST00000433696.6
ENST00000538969.5 ENST00000544133.5 |
CD36
|
CD36 molecule |
| chr22_+_19760714 | 1.66 |
ENST00000649276.2
|
TBX1
|
T-box transcription factor 1 |
| chr11_-_86955385 | 1.61 |
ENST00000531380.2
|
FZD4
|
frizzled class receptor 4 |
| chr7_+_80638633 | 1.58 |
ENST00000447544.7
ENST00000482059.6 |
CD36
|
CD36 molecule |
| chr7_+_80638662 | 1.57 |
ENST00000394788.7
|
CD36
|
CD36 molecule |
| chr13_+_110305806 | 1.54 |
ENST00000400163.7
|
COL4A2
|
collagen type IV alpha 2 chain |
| chr18_+_6729698 | 1.46 |
ENST00000383472.9
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr2_-_1744442 | 1.38 |
ENST00000433670.5
ENST00000425171.1 ENST00000252804.9 |
PXDN
|
peroxidasin |
| chr12_+_53097656 | 1.38 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr7_-_131556602 | 1.37 |
ENST00000322985.9
ENST00000378555.8 |
PODXL
|
podocalyxin like |
| chr11_+_114060204 | 1.33 |
ENST00000683318.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr12_+_54854505 | 1.25 |
ENST00000308796.11
ENST00000619042.1 |
MUCL1
|
mucin like 1 |
| chr7_-_108003122 | 1.16 |
ENST00000393559.2
ENST00000222399.11 ENST00000676777.1 ENST00000439976.6 ENST00000393560.5 ENST00000677793.1 ENST00000679244.1 |
LAMB1
|
laminin subunit beta 1 |
| chr14_+_75280078 | 1.16 |
ENST00000555347.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr19_-_18606779 | 1.15 |
ENST00000684169.1
ENST00000392386.8 |
CRLF1
|
cytokine receptor like factor 1 |
| chr5_-_39425187 | 1.14 |
ENST00000545653.5
|
DAB2
|
DAB adaptor protein 2 |
| chr9_+_110090197 | 1.12 |
ENST00000480388.1
|
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chr20_+_32010429 | 1.07 |
ENST00000452892.3
ENST00000262659.12 |
CCM2L
|
CCM2 like scaffold protein |
| chr10_+_24466487 | 1.06 |
ENST00000396446.5
ENST00000396445.5 ENST00000376451.4 |
KIAA1217
|
KIAA1217 |
| chr20_-_62367304 | 1.03 |
ENST00000252999.7
|
LAMA5
|
laminin subunit alpha 5 |
| chr4_+_77157189 | 1.03 |
ENST00000316355.10
ENST00000502280.5 |
CCNG2
|
cyclin G2 |
| chr12_+_101666203 | 1.02 |
ENST00000549608.1
|
MYBPC1
|
myosin binding protein C1 |
| chr3_-_134373719 | 0.98 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr5_+_102865244 | 0.94 |
ENST00000511477.5
ENST00000506006.1 ENST00000509832.5 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr8_-_6563238 | 0.91 |
ENST00000629816.3
ENST00000523120.2 |
ANGPT2
|
angiopoietin 2 |
| chr18_+_3450036 | 0.91 |
ENST00000546979.5
ENST00000343820.10 ENST00000551402.1 ENST00000577543.5 |
TGIF1
|
TGFB induced factor homeobox 1 |
| chr12_-_10098977 | 0.86 |
ENST00000315330.8
ENST00000457018.6 |
CLEC1A
|
C-type lectin domain family 1 member A |
| chr16_+_54930827 | 0.85 |
ENST00000394636.9
|
IRX5
|
iroquois homeobox 5 |
| chr19_+_55600277 | 0.84 |
ENST00000301073.4
|
ZNF524
|
zinc finger protein 524 |
| chr12_-_10098940 | 0.84 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1 member A |
| chr6_-_123636923 | 0.81 |
ENST00000334268.9
|
TRDN
|
triadin |
| chr6_-_123636979 | 0.81 |
ENST00000662930.1
|
TRDN
|
triadin |
| chr6_-_75206044 | 0.80 |
ENST00000322507.13
|
COL12A1
|
collagen type XII alpha 1 chain |
| chr6_-_106974721 | 0.80 |
ENST00000606017.2
ENST00000620405.1 |
CD24
|
CD24 molecule |
| chr8_-_6563044 | 0.80 |
ENST00000338312.10
|
ANGPT2
|
angiopoietin 2 |
| chr8_-_6563409 | 0.79 |
ENST00000325203.9
|
ANGPT2
|
angiopoietin 2 |
| chr5_-_39424966 | 0.79 |
ENST00000515700.5
ENST00000320816.11 ENST00000339788.10 |
DAB2
|
DAB adaptor protein 2 |
| chr7_+_116672187 | 0.76 |
ENST00000318493.11
ENST00000397752.8 |
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr19_-_54257217 | 0.74 |
ENST00000345866.10
ENST00000449561.3 |
LILRB5
|
leukocyte immunoglobulin like receptor B5 |
| chr19_-_17264718 | 0.74 |
ENST00000431146.6
ENST00000594190.5 |
USHBP1
|
USH1 protein network component harmonin binding protein 1 |
| chr3_-_186544377 | 0.72 |
ENST00000307944.6
|
CRYGS
|
crystallin gamma S |
| chr18_+_7946841 | 0.71 |
ENST00000578916.2
|
PTPRM
|
protein tyrosine phosphatase receptor type M |
| chr19_-_54257285 | 0.71 |
ENST00000316219.9
|
LILRB5
|
leukocyte immunoglobulin like receptor B5 |
| chr7_-_93226449 | 0.68 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr2_+_222671651 | 0.67 |
ENST00000446656.4
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1 |
| chr2_+_54457190 | 0.67 |
ENST00000389980.7
|
SPTBN1
|
spectrin beta, non-erythrocytic 1 |
| chr14_+_75279961 | 0.67 |
ENST00000557139.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr18_+_3449817 | 0.65 |
ENST00000407501.6
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr9_-_14313843 | 0.63 |
ENST00000636063.1
ENST00000380921.3 ENST00000622520.1 ENST00000380959.7 |
NFIB
|
nuclear factor I B |
| chr4_+_101813810 | 0.63 |
ENST00000444316.2
|
BANK1
|
B cell scaffold protein with ankyrin repeats 1 |
| chr1_+_67685170 | 0.63 |
ENST00000370985.4
ENST00000370986.9 ENST00000650283.1 ENST00000648742.1 |
GADD45A
|
growth arrest and DNA damage inducible alpha |
| chr9_-_72364504 | 0.61 |
ENST00000237937.7
ENST00000343431.6 ENST00000376956.3 |
ZFAND5
|
zinc finger AN1-type containing 5 |
| chr5_+_141182369 | 0.61 |
ENST00000609684.3
ENST00000625044.1 ENST00000623407.1 ENST00000623884.1 |
PCDHB16
ENSG00000279068.1
|
protocadherin beta 16 novel transcript |
| chr10_-_104085847 | 0.60 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr1_-_93180261 | 0.59 |
ENST00000370280.1
ENST00000479918.5 |
TMED5
|
transmembrane p24 trafficking protein 5 |
| chr6_-_123636997 | 0.59 |
ENST00000546248.5
|
TRDN
|
triadin |
| chr17_+_69502397 | 0.59 |
ENST00000613873.4
ENST00000589647.5 |
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr11_-_102843597 | 0.58 |
ENST00000299855.10
|
MMP3
|
matrix metallopeptidase 3 |
| chr2_+_69013170 | 0.58 |
ENST00000303714.9
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr2_-_85414039 | 0.57 |
ENST00000447219.6
ENST00000409670.5 ENST00000409724.5 |
CAPG
|
capping actin protein, gelsolin like |
| chr20_-_40689228 | 0.56 |
ENST00000373313.3
|
MAFB
|
MAF bZIP transcription factor B |
| chr20_-_57709760 | 0.54 |
ENST00000395819.3
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr1_+_93448155 | 0.54 |
ENST00000370253.6
|
FNBP1L
|
formin binding protein 1 like |
| chr19_-_17264732 | 0.54 |
ENST00000252597.8
|
USHBP1
|
USH1 protein network component harmonin binding protein 1 |
| chr16_-_73048104 | 0.52 |
ENST00000268489.10
|
ZFHX3
|
zinc finger homeobox 3 |
| chr17_-_10469558 | 0.52 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4 |
| chr7_-_140176970 | 0.51 |
ENST00000397560.7
|
KDM7A
|
lysine demethylase 7A |
| chr5_+_102865805 | 0.51 |
ENST00000346918.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_+_201739864 | 0.51 |
ENST00000367295.5
|
NAV1
|
neuron navigator 1 |
| chr3_-_50611767 | 0.51 |
ENST00000443053.6
ENST00000348721.4 |
CISH
|
cytokine inducible SH2 containing protein |
| chr15_+_90872862 | 0.50 |
ENST00000618099.4
|
FURIN
|
furin, paired basic amino acid cleaving enzyme |
| chr16_+_3018390 | 0.50 |
ENST00000573001.5
|
TNFRSF12A
|
TNF receptor superfamily member 12A |
| chr6_-_123636906 | 0.50 |
ENST00000628709.2
|
TRDN
|
triadin |
| chr2_+_201116940 | 0.50 |
ENST00000433445.1
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr10_-_104085793 | 0.49 |
ENST00000650263.1
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr1_+_185734362 | 0.49 |
ENST00000271588.9
|
HMCN1
|
hemicentin 1 |
| chr14_-_73027077 | 0.49 |
ENST00000553891.5
ENST00000556143.6 |
ZFYVE1
|
zinc finger FYVE-type containing 1 |
| chr6_-_106975309 | 0.49 |
ENST00000615659.1
|
CD24
|
CD24 molecule |
| chr14_-_73027117 | 0.48 |
ENST00000318876.9
|
ZFYVE1
|
zinc finger FYVE-type containing 1 |
| chr6_-_166627244 | 0.48 |
ENST00000265678.9
|
RPS6KA2
|
ribosomal protein S6 kinase A2 |
| chr2_+_36696758 | 0.47 |
ENST00000457137.6
|
VIT
|
vitrin |
| chr18_-_80247348 | 0.47 |
ENST00000470488.2
ENST00000353265.8 |
PARD6G
|
par-6 family cell polarity regulator gamma |
| chr20_+_41136944 | 0.46 |
ENST00000244007.7
|
PLCG1
|
phospholipase C gamma 1 |
| chr8_-_16186270 | 0.46 |
ENST00000445506.6
|
MSR1
|
macrophage scavenger receptor 1 |
| chr1_+_15758768 | 0.46 |
ENST00000483633.6
ENST00000502739.5 ENST00000375766.8 ENST00000431771.6 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr20_-_13990609 | 0.46 |
ENST00000284951.10
ENST00000378072.5 |
SEL1L2
|
SEL1L2 adaptor subunit of ERAD E3 ligase |
| chr9_-_71768386 | 0.45 |
ENST00000377066.9
ENST00000377044.9 |
CEMIP2
|
cell migration inducing hyaluronidase 2 |
| chr22_-_31292445 | 0.44 |
ENST00000402249.7
ENST00000215912.10 ENST00000443175.1 ENST00000441972.5 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr11_-_5254741 | 0.44 |
ENST00000444587.1
ENST00000336906.6 ENST00000642908.1 ENST00000647543.1 |
HBG2
ENSG00000284931.1
|
hemoglobin subunit gamma 2 novel protein |
| chr5_-_39424859 | 0.43 |
ENST00000503513.5
|
DAB2
|
DAB adaptor protein 2 |
| chr6_+_20403679 | 0.43 |
ENST00000535432.2
|
E2F3
|
E2F transcription factor 3 |
| chr10_-_104085762 | 0.43 |
ENST00000393211.3
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr12_+_95217792 | 0.43 |
ENST00000436874.6
ENST00000551472.5 ENST00000552821.5 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr17_+_17042505 | 0.42 |
ENST00000577514.5
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr15_-_72272530 | 0.41 |
ENST00000569795.6
ENST00000566844.1 |
PARP6
|
poly(ADP-ribose) polymerase family member 6 |
| chr5_+_102865737 | 0.41 |
ENST00000509523.2
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr11_-_10808913 | 0.41 |
ENST00000527419.6
ENST00000530211.6 ENST00000339995.11 ENST00000530702.2 ENST00000524932.6 ENST00000532570.6 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma 2 |
| chr8_-_17722217 | 0.40 |
ENST00000381861.7
|
MTUS1
|
microtubule associated scaffold protein 1 |
| chr5_+_141100466 | 0.40 |
ENST00000231130.3
|
PCDHB3
|
protocadherin beta 3 |
| chr14_-_31207469 | 0.40 |
ENST00000556224.5
|
HECTD1
|
HECT domain E3 ubiquitin protein ligase 1 |
| chr1_-_225653045 | 0.40 |
ENST00000366843.6
ENST00000366844.7 |
ENAH
|
ENAH actin regulator |
| chr1_+_244835616 | 0.39 |
ENST00000366528.3
ENST00000411948.7 |
COX20
|
cytochrome c oxidase assembly factor COX20 |
| chr1_+_27934980 | 0.38 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr12_-_109477293 | 0.38 |
ENST00000228495.11
ENST00000542858.1 ENST00000542262.5 |
KCTD10
|
potassium channel tetramerization domain containing 10 |
| chr7_+_116672357 | 0.38 |
ENST00000456159.1
|
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr7_+_16753731 | 0.38 |
ENST00000262067.5
|
TSPAN13
|
tetraspanin 13 |
| chr20_-_35954461 | 0.38 |
ENST00000305978.7
|
SCAND1
|
SCAN domain containing 1 |
| chr1_+_27935022 | 0.37 |
ENST00000411604.5
ENST00000373888.8 |
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr13_+_111203052 | 0.37 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor 7 |
| chr7_+_30145789 | 0.37 |
ENST00000324489.5
|
MTURN
|
maturin, neural progenitor differentiation regulator homolog |
| chr22_+_31081310 | 0.37 |
ENST00000426927.5
ENST00000482444.5 ENST00000440425.5 ENST00000333137.12 ENST00000358743.5 ENST00000347557.6 |
SMTN
|
smoothelin |
| chr12_+_95217760 | 0.37 |
ENST00000549002.5
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr7_-_84492718 | 0.37 |
ENST00000424555.5
|
SEMA3A
|
semaphorin 3A |
| chr12_+_8697177 | 0.37 |
ENST00000541044.5
|
RIMKLB
|
ribosomal modification protein rimK like family member B |
| chr15_-_70097874 | 0.36 |
ENST00000557997.5
ENST00000317509.12 ENST00000451782.7 ENST00000627388.2 |
TLE3
|
TLE family member 3, transcriptional corepressor |
| chr17_-_76570544 | 0.35 |
ENST00000640006.1
|
ENSG00000284526.1
|
novel protein |
| chr2_+_36696790 | 0.34 |
ENST00000497382.5
ENST00000404084.5 ENST00000379241.7 ENST00000401530.5 |
VIT
|
vitrin |
| chr8_-_143986425 | 0.33 |
ENST00000313059.9
ENST00000524918.5 ENST00000313028.12 ENST00000525773.5 |
PARP10
|
poly(ADP-ribose) polymerase family member 10 |
| chr3_+_50611871 | 0.33 |
ENST00000446044.5
|
MAPKAPK3
|
MAPK activated protein kinase 3 |
| chr1_+_27935110 | 0.33 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr11_-_10808304 | 0.33 |
ENST00000532082.6
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma 2 |
| chr5_-_173616588 | 0.32 |
ENST00000285908.5
ENST00000311086.9 ENST00000480951.1 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr15_-_40874216 | 0.32 |
ENST00000220507.5
|
RHOV
|
ras homolog family member V |
| chr6_+_139135063 | 0.32 |
ENST00000367658.3
|
HECA
|
hdc homolog, cell cycle regulator |
| chr5_+_110738134 | 0.32 |
ENST00000513807.5
|
SLC25A46
|
solute carrier family 25 member 46 |
| chr2_-_20225433 | 0.31 |
ENST00000381150.5
|
SDC1
|
syndecan 1 |
| chr4_+_77158252 | 0.31 |
ENST00000395640.5
|
CCNG2
|
cyclin G2 |
| chr4_+_26321365 | 0.31 |
ENST00000505958.6
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr4_+_77158356 | 0.31 |
ENST00000512918.5
|
CCNG2
|
cyclin G2 |
| chr7_-_27180013 | 0.31 |
ENST00000470747.4
|
ENSG00000257184.3
|
HOXA10-HOXA9 readthrough |
| chr4_+_70397931 | 0.30 |
ENST00000399575.7
|
OPRPN
|
opiorphin prepropeptide |
| chr5_-_136365476 | 0.30 |
ENST00000378459.7
ENST00000502753.4 ENST00000513104.6 ENST00000352189.8 |
TRPC7
|
transient receptor potential cation channel subfamily C member 7 |
| chr6_-_123636966 | 0.30 |
ENST00000542443.5
|
TRDN
|
triadin |
| chrX_-_129843388 | 0.30 |
ENST00000371064.7
|
ZDHHC9
|
zinc finger DHHC-type palmitoyltransferase 9 |
| chr1_-_153967621 | 0.30 |
ENST00000413622.5
ENST00000310483.10 |
SLC39A1
|
solute carrier family 39 member 1 |
| chr16_+_474917 | 0.30 |
ENST00000449879.6
|
RAB11FIP3
|
RAB11 family interacting protein 3 |
| chr15_+_81000913 | 0.30 |
ENST00000267984.4
|
TLNRD1
|
talin rod domain containing 1 |
| chr7_-_111392915 | 0.29 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr4_+_26321192 | 0.29 |
ENST00000681484.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr13_-_41019289 | 0.29 |
ENST00000239882.7
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr7_-_84492547 | 0.28 |
ENST00000448879.5
|
SEMA3A
|
semaphorin 3A |
| chr9_-_101435760 | 0.28 |
ENST00000647789.2
ENST00000616752.1 |
ALDOB
|
aldolase, fructose-bisphosphate B |
| chr5_+_140855882 | 0.28 |
ENST00000562220.2
ENST00000307360.6 ENST00000506939.6 |
PCDHA10
|
protocadherin alpha 10 |
| chr11_-_70717994 | 0.28 |
ENST00000659264.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr16_+_474850 | 0.28 |
ENST00000450428.5
ENST00000452814.5 |
RAB11FIP3
|
RAB11 family interacting protein 3 |
| chr9_-_72365198 | 0.27 |
ENST00000376962.10
ENST00000376960.8 |
ZFAND5
|
zinc finger AN1-type containing 5 |
| chr16_-_58629852 | 0.27 |
ENST00000569020.5
|
CNOT1
|
CCR4-NOT transcription complex subunit 1 |
| chr7_+_116210501 | 0.27 |
ENST00000455989.1
ENST00000358204.9 |
TES
|
testin LIM domain protein |
| chr14_-_88554898 | 0.27 |
ENST00000556564.6
|
PTPN21
|
protein tyrosine phosphatase non-receptor type 21 |
| chr16_+_21597204 | 0.27 |
ENST00000567404.5
|
METTL9
|
methyltransferase like 9 |
| chr3_+_109136707 | 0.27 |
ENST00000622536.6
|
C3orf85
|
chromosome 3 open reading frame 85 |
| chr1_+_93179883 | 0.25 |
ENST00000343253.11
|
CCDC18
|
coiled-coil domain containing 18 |
| chr17_+_69502154 | 0.25 |
ENST00000589295.5
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr5_-_176537361 | 0.25 |
ENST00000274811.9
|
RNF44
|
ring finger protein 44 |
| chr2_-_37671633 | 0.25 |
ENST00000295324.4
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr2_+_36696686 | 0.25 |
ENST00000379242.7
ENST00000389975.7 |
VIT
|
vitrin |
| chr6_-_41072529 | 0.24 |
ENST00000373154.6
ENST00000464633.5 ENST00000628419.2 ENST00000479950.5 ENST00000482515.5 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr12_+_93569814 | 0.24 |
ENST00000340600.6
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr4_+_2818155 | 0.24 |
ENST00000511747.6
|
SH3BP2
|
SH3 domain binding protein 2 |
| chr17_+_17042433 | 0.23 |
ENST00000651222.2
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr3_+_179148341 | 0.23 |
ENST00000263967.4
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr6_-_168067509 | 0.23 |
ENST00000336070.11
|
FRMD1
|
FERM domain containing 1 |
| chr7_+_141708353 | 0.23 |
ENST00000397541.6
|
WEE2
|
WEE2 oocyte meiosis inhibiting kinase |
| chr1_-_152325232 | 0.22 |
ENST00000368799.2
|
FLG
|
filaggrin |
| chr15_-_70097852 | 0.22 |
ENST00000559191.5
|
TLE3
|
TLE family member 3, transcriptional corepressor |
| chr6_+_10555787 | 0.22 |
ENST00000316170.9
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr7_-_149028651 | 0.22 |
ENST00000286091.9
|
PDIA4
|
protein disulfide isomerase family A member 4 |
| chr12_-_76423256 | 0.22 |
ENST00000546946.5
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr5_+_72816643 | 0.21 |
ENST00000337273.10
ENST00000523768.5 |
TNPO1
|
transportin 1 |
| chr4_-_183322426 | 0.21 |
ENST00000541814.1
|
CLDN24
|
claudin 24 |
| chr2_-_37672448 | 0.20 |
ENST00000611976.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr15_-_93089192 | 0.20 |
ENST00000329082.11
|
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chrX_-_136880754 | 0.20 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein X-linked |
| chr12_-_76486061 | 0.20 |
ENST00000548341.5
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr16_-_57846535 | 0.20 |
ENST00000565684.5
|
KIFC3
|
kinesin family member C3 |
| chr12_+_68610858 | 0.20 |
ENST00000393436.9
ENST00000425247.6 ENST00000250559.14 ENST00000489473.6 ENST00000422358.6 ENST00000541167.5 ENST00000538283.5 ENST00000341355.9 ENST00000537460.5 ENST00000450214.6 ENST00000545270.5 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr14_+_35292429 | 0.19 |
ENST00000555764.5
ENST00000556506.1 |
PSMA6
|
proteasome 20S subunit alpha 6 |
| chrX_-_111410919 | 0.19 |
ENST00000496551.2
|
DCX
|
doublecortin |
| chr20_+_35954564 | 0.19 |
ENST00000622112.4
ENST00000614708.1 |
CNBD2
|
cyclic nucleotide binding domain containing 2 |
| chr4_-_169270849 | 0.19 |
ENST00000502315.1
ENST00000284637.14 |
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr17_+_59565598 | 0.19 |
ENST00000251241.9
ENST00000425628.7 ENST00000584385.5 ENST00000580030.1 |
DHX40
|
DEAH-box helicase 40 |
| chr7_-_149028452 | 0.19 |
ENST00000413966.1
ENST00000652332.1 |
PDIA4
|
protein disulfide isomerase family A member 4 |
| chr19_-_8149586 | 0.19 |
ENST00000600128.6
|
FBN3
|
fibrillin 3 |
| chr8_+_10054269 | 0.19 |
ENST00000317173.9
ENST00000441698.6 |
MSRA
|
methionine sulfoxide reductase A |
| chr3_-_128488507 | 0.18 |
ENST00000487848.5
|
GATA2
|
GATA binding protein 2 |
| chrX_+_12975216 | 0.18 |
ENST00000380635.5
|
TMSB4X
|
thymosin beta 4 X-linked |
| chr7_+_134779663 | 0.18 |
ENST00000361901.6
|
CALD1
|
caldesmon 1 |
| chr18_+_58045642 | 0.18 |
ENST00000676223.1
ENST00000675147.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr7_+_100101657 | 0.18 |
ENST00000421755.5
|
AP4M1
|
adaptor related protein complex 4 subunit mu 1 |
| chr12_+_68610918 | 0.17 |
ENST00000538980.5
ENST00000542018.5 ENST00000543393.5 ENST00000534899.5 ENST00000453560.6 ENST00000378985.7 ENST00000540209.5 ENST00000540781.5 ENST00000535492.5 ENST00000539091.5 ENST00000542145.5 ENST00000485252.6 ENST00000541386.5 ENST00000538877.5 ENST00000543697.5 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr20_+_38066166 | 0.17 |
ENST00000622494.1
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr7_+_100101556 | 0.17 |
ENST00000438383.5
ENST00000429084.5 ENST00000439416.5 |
AP4M1
|
adaptor related protein complex 4 subunit mu 1 |
| chr13_+_39038347 | 0.17 |
ENST00000379599.6
ENST00000379600.8 |
NHLRC3
|
NHL repeat containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.3 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.8 | 2.5 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.6 | 4.5 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.6 | 3.0 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 0.6 | 1.7 | GO:0035981 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.5 | 1.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.5 | 2.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.4 | 4.8 | GO:2000334 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.4 | 1.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.3 | 1.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 1.3 | GO:0032595 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.3 | 1.9 | GO:0018032 | protein amidation(GO:0018032) |
| 0.2 | 1.3 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.2 | 0.9 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 1.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 1.8 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.2 | 1.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.6 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.9 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.6 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.7 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.1 | 0.5 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 0.7 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.5 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 1.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.3 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.3 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 0.2 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 0.2 | GO:0097698 | telomere maintenance via base-excision repair(GO:0097698) |
| 0.1 | 1.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.3 | GO:0022018 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.1 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.6 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.6 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.6 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 1.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 2.0 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.5 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.4 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 1.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.5 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.5 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 3.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.3 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.6 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.6 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.2 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 1.0 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 1.0 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.7 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 1.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 1.4 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 1.0 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 1.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.4 | 6.1 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.3 | 2.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 1.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 1.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 3.0 | GO:0030314 | junctional membrane complex(GO:0030314) sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 1.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 9.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 0.8 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 1.4 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.2 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.1 | 0.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 1.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 1.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.5 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 1.5 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.7 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 2.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.3 | 2.7 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 1.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 1.9 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.2 | 0.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 1.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 0.5 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.1 | 1.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.5 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 1.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.7 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 2.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.3 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.1 | 1.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.2 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.8 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 1.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 4.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 2.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 1.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 2.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 2.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 3.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 3.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 2.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 3.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 2.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 8.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 5.0 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 1.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.1 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 2.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 2.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |