|
chr14_-_23435652
Show fit
|
10.06 |
ENST00000355349.4
|
MYH7
|
myosin heavy chain 7
|
|
chr12_-_110920568
Show fit
|
5.62 |
ENST00000548438.1
ENST00000228841.15
|
MYL2
|
myosin light chain 2
|
|
chr17_-_10549612
Show fit
|
4.79 |
ENST00000532183.6
ENST00000397183.6
ENST00000420805.1
|
MYH2
|
myosin heavy chain 2
|
|
chr17_-_10549652
Show fit
|
4.34 |
ENST00000245503.10
|
MYH2
|
myosin heavy chain 2
|
|
chrX_+_136169624
Show fit
|
3.20 |
ENST00000394153.6
|
FHL1
|
four and a half LIM domains 1
|
|
chr12_-_6124662
Show fit
|
2.80 |
ENST00000261405.10
|
VWF
|
von Willebrand factor
|
|
chrX_+_136169664
Show fit
|
2.78 |
ENST00000456445.5
|
FHL1
|
four and a half LIM domains 1
|
|
chrX_+_136169891
Show fit
|
2.74 |
ENST00000449474.5
|
FHL1
|
four and a half LIM domains 1
|
|
chrX_+_136169833
Show fit
|
2.73 |
ENST00000628032.2
|
FHL1
|
four and a half LIM domains 1
|
|
chr12_-_110920710
Show fit
|
2.67 |
ENST00000546404.1
|
MYL2
|
myosin light chain 2
|
|
chr17_+_4948252
Show fit
|
2.59 |
ENST00000520221.5
|
ENO3
|
enolase 3
|
|
chr17_-_10549694
Show fit
|
2.59 |
ENST00000622564.4
|
MYH2
|
myosin heavy chain 2
|
|
chr6_-_46080332
Show fit
|
2.31 |
ENST00000185206.12
|
CLIC5
|
chloride intracellular channel 5
|
|
chr12_-_8662808
Show fit
|
2.30 |
ENST00000359478.7
ENST00000396549.6
|
MFAP5
|
microfibril associated protein 5
|
|
chr10_+_68106109
Show fit
|
2.27 |
ENST00000540630.5
ENST00000354393.6
|
MYPN
|
myopalladin
|
|
chr3_-_52452828
Show fit
|
2.21 |
ENST00000496590.1
|
TNNC1
|
troponin C1, slow skeletal and cardiac type
|
|
chr6_-_139291987
Show fit
|
2.15 |
ENST00000358430.8
|
TXLNB
|
taxilin beta
|
|
chr9_-_35684766
Show fit
|
2.14 |
ENST00000644325.1
|
TPM2
|
tropomyosin 2
|
|
chr3_-_46863435
Show fit
|
2.09 |
ENST00000395869.5
ENST00000653454.1
ENST00000292327.6
|
MYL3
|
myosin light chain 3
|
|
chr20_+_17227020
Show fit
|
2.08 |
ENST00000262545.7
ENST00000536609.1
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr15_+_49423233
Show fit
|
1.98 |
ENST00000560270.1
ENST00000267843.9
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7
|
|
chr5_-_139444470
Show fit
|
1.77 |
ENST00000512473.5
ENST00000515581.5
ENST00000515277.5
|
DNAJC18
|
DnaJ heat shock protein family (Hsp40) member C18
|
|
chr10_-_114684612
Show fit
|
1.73 |
ENST00000533213.6
ENST00000369252.8
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr10_-_114685000
Show fit
|
1.70 |
ENST00000369256.6
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr18_+_58341038
Show fit
|
1.53 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr12_-_8662881
Show fit
|
1.50 |
ENST00000433590.6
|
MFAP5
|
microfibril associated protein 5
|
|
chr13_+_110305806
Show fit
|
1.49 |
ENST00000400163.7
|
COL4A2
|
collagen type IV alpha 2 chain
|
|
chr4_-_64409444
Show fit
|
1.48 |
ENST00000381210.8
ENST00000507440.5
|
TECRL
|
trans-2,3-enoyl-CoA reductase like
|
|
chr12_-_16605939
Show fit
|
1.45 |
ENST00000541295.5
ENST00000535535.5
|
LMO3
|
LIM domain only 3
|
|
chr5_-_88824334
Show fit
|
1.41 |
ENST00000506716.5
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr6_+_12290353
Show fit
|
1.38 |
ENST00000379375.6
|
EDN1
|
endothelin 1
|
|
chr10_+_24239181
Show fit
|
1.38 |
ENST00000438429.5
|
KIAA1217
|
KIAA1217
|
|
chr14_-_91947654
Show fit
|
1.33 |
ENST00000342058.9
|
FBLN5
|
fibulin 5
|
|
chr10_+_11164961
Show fit
|
1.30 |
ENST00000399850.7
ENST00000417956.6
|
CELF2
|
CUGBP Elav-like family member 2
|
|
chr16_-_11281322
Show fit
|
1.27 |
ENST00000312511.4
|
PRM1
|
protamine 1
|
|
chr11_+_112961247
Show fit
|
1.26 |
ENST00000621518.4
ENST00000618266.4
ENST00000615112.4
ENST00000615285.4
ENST00000619839.4
ENST00000401611.6
ENST00000621128.4
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr12_+_59664677
Show fit
|
1.22 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7
|
|
chr12_-_16606102
Show fit
|
1.21 |
ENST00000537304.6
|
LMO3
|
LIM domain only 3
|
|
chr4_-_185810894
Show fit
|
1.18 |
ENST00000448662.6
ENST00000439049.5
ENST00000420158.5
ENST00000319471.13
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr18_+_34818436
Show fit
|
1.18 |
ENST00000599844.5
ENST00000679731.1
|
DTNA
|
dystrobrevin alpha
|
|
chr3_+_189631373
Show fit
|
1.16 |
ENST00000264731.8
ENST00000418709.6
ENST00000320472.9
ENST00000392460.7
ENST00000440651.6
|
TP63
|
tumor protein p63
|
|
chr5_-_159099909
Show fit
|
1.16 |
ENST00000313708.11
|
EBF1
|
EBF transcription factor 1
|
|
chr3_-_73433904
Show fit
|
1.14 |
ENST00000479530.5
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr6_+_122399621
Show fit
|
1.14 |
ENST00000368455.9
|
HSF2
|
heat shock transcription factor 2
|
|
chr4_+_41612892
Show fit
|
1.12 |
ENST00000509454.5
ENST00000396595.7
ENST00000381753.8
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr3_+_189789734
Show fit
|
1.12 |
ENST00000437221.5
ENST00000392463.6
ENST00000392461.7
ENST00000449992.5
ENST00000456148.1
|
TP63
|
tumor protein p63
|
|
chr2_+_69013170
Show fit
|
1.11 |
ENST00000303714.9
|
ANTXR1
|
ANTXR cell adhesion molecule 1
|
|
chr1_-_151146611
Show fit
|
1.11 |
ENST00000341697.7
ENST00000368914.8
|
SEMA6C
|
semaphorin 6C
|
|
chr1_-_36440873
Show fit
|
1.11 |
ENST00000433045.6
|
OSCP1
|
organic solute carrier partner 1
|
|
chr2_+_69013414
Show fit
|
1.09 |
ENST00000681816.1
ENST00000482235.2
|
ANTXR1
|
ANTXR cell adhesion molecule 1
|
|
chr6_+_122399536
Show fit
|
1.09 |
ENST00000452194.5
|
HSF2
|
heat shock transcription factor 2
|
|
chr14_-_91947383
Show fit
|
1.06 |
ENST00000267620.14
|
FBLN5
|
fibulin 5
|
|
chr1_+_3021466
Show fit
|
1.05 |
ENST00000378404.4
|
ACTRT2
|
actin related protein T2
|
|
chr10_+_50990864
Show fit
|
1.03 |
ENST00000401604.8
|
PRKG1
|
protein kinase cGMP-dependent 1
|
|
chr3_+_89107613
Show fit
|
1.01 |
ENST00000336596.7
|
EPHA3
|
EPH receptor A3
|
|
chr11_+_112961402
Show fit
|
0.99 |
ENST00000613217.4
ENST00000316851.12
ENST00000620046.4
ENST00000531044.5
ENST00000529356.5
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr2_+_69013379
Show fit
|
0.98 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1
|
|
chr8_-_40897814
Show fit
|
0.98 |
ENST00000297737.11
ENST00000315769.11
|
ZMAT4
|
zinc finger matrin-type 4
|
|
chr14_-_91947950
Show fit
|
0.95 |
ENST00000554468.5
|
FBLN5
|
fibulin 5
|
|
chr5_-_88824266
Show fit
|
0.95 |
ENST00000509373.1
ENST00000636541.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr8_+_143213192
Show fit
|
0.95 |
ENST00000622500.2
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1
|
|
chr17_-_64390852
Show fit
|
0.94 |
ENST00000563924.6
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1
|
|
chr3_+_189789643
Show fit
|
0.94 |
ENST00000354600.10
|
TP63
|
tumor protein p63
|
|
chr18_+_58864866
Show fit
|
0.92 |
ENST00000588456.5
ENST00000591808.6
ENST00000589481.1
ENST00000591049.1
|
ZNF532
|
zinc finger protein 532
|
|
chr4_-_64409381
Show fit
|
0.92 |
ENST00000509536.1
|
TECRL
|
trans-2,3-enoyl-CoA reductase like
|
|
chr11_+_131911396
Show fit
|
0.89 |
ENST00000425719.6
ENST00000374784.5
|
NTM
|
neurotrimin
|
|
chr8_-_107497909
Show fit
|
0.89 |
ENST00000517746.6
|
ANGPT1
|
angiopoietin 1
|
|
chr15_-_48963912
Show fit
|
0.87 |
ENST00000332408.9
|
SHC4
|
SHC adaptor protein 4
|
|
chr1_-_151146643
Show fit
|
0.87 |
ENST00000613223.1
|
SEMA6C
|
semaphorin 6C
|
|
chr12_-_85836372
Show fit
|
0.86 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9
|
|
chr5_-_168883333
Show fit
|
0.85 |
ENST00000404867.7
|
SLIT3
|
slit guidance ligand 3
|
|
chr17_-_41586887
Show fit
|
0.84 |
ENST00000167586.7
|
KRT14
|
keratin 14
|
|
chr1_+_164559766
Show fit
|
0.83 |
ENST00000367897.5
ENST00000559240.5
|
PBX1
|
PBX homeobox 1
|
|
chr8_+_67952028
Show fit
|
0.83 |
ENST00000288368.5
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 2
|
|
chrX_-_112840815
Show fit
|
0.83 |
ENST00000304758.5
ENST00000371959.9
|
AMOT
|
angiomotin
|
|
chr4_-_100517991
Show fit
|
0.82 |
ENST00000511970.5
ENST00000502569.1
ENST00000305864.7
ENST00000296420.9
|
EMCN
|
endomucin
|
|
chrX_+_15507302
Show fit
|
0.82 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr14_-_91946989
Show fit
|
0.81 |
ENST00000556154.5
|
FBLN5
|
fibulin 5
|
|
chr22_-_35840218
Show fit
|
0.80 |
ENST00000414461.6
ENST00000416721.6
ENST00000449924.6
ENST00000262829.11
ENST00000397305.3
|
RBFOX2
|
RNA binding fox-1 homolog 2
|
|
chr12_+_53050179
Show fit
|
0.79 |
ENST00000546602.5
ENST00000552570.5
ENST00000549700.5
|
TNS2
|
tensin 2
|
|
chr9_-_120477354
Show fit
|
0.79 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2
|
|
chr1_+_164559739
Show fit
|
0.78 |
ENST00000627490.2
|
PBX1
|
PBX homeobox 1
|
|
chr4_+_41612702
Show fit
|
0.78 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr17_-_64390592
Show fit
|
0.77 |
ENST00000563523.5
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1
|
|
chr6_+_148508446
Show fit
|
0.76 |
ENST00000637729.1
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr8_+_102551583
Show fit
|
0.76 |
ENST00000285402.4
|
ODF1
|
outer dense fiber of sperm tails 1
|
|
chr8_-_42501224
Show fit
|
0.75 |
ENST00000520262.6
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 member 2
|
|
chr4_-_23881282
Show fit
|
0.74 |
ENST00000613098.4
|
PPARGC1A
|
PPARG coactivator 1 alpha
|
|
chrX_-_13817027
Show fit
|
0.74 |
ENST00000493677.5
ENST00000355135.6
ENST00000316715.9
|
GPM6B
|
glycoprotein M6B
|
|
chr3_+_89107649
Show fit
|
0.74 |
ENST00000452448.6
ENST00000494014.1
|
EPHA3
|
EPH receptor A3
|
|
chr10_+_61901678
Show fit
|
0.74 |
ENST00000644638.1
ENST00000681100.1
ENST00000279873.12
|
ARID5B
|
AT-rich interaction domain 5B
|
|
chr19_-_7633713
Show fit
|
0.74 |
ENST00000311069.6
|
PCP2
|
Purkinje cell protein 2
|
|
chr12_-_102480552
Show fit
|
0.73 |
ENST00000337514.11
ENST00000307046.8
|
IGF1
|
insulin like growth factor 1
|
|
chr21_-_38498415
Show fit
|
0.73 |
ENST00000398905.5
ENST00000398907.5
ENST00000453032.6
ENST00000288319.12
|
ERG
|
ETS transcription factor ERG
|
|
chr9_+_126335200
Show fit
|
0.73 |
ENST00000402437.2
|
MVB12B
|
multivesicular body subunit 12B
|
|
chr2_-_187448244
Show fit
|
0.72 |
ENST00000392370.8
ENST00000410068.5
ENST00000447403.5
ENST00000410102.5
|
CALCRL
|
calcitonin receptor like receptor
|
|
chr7_-_16881967
Show fit
|
0.72 |
ENST00000402239.7
ENST00000310398.7
ENST00000414935.1
|
AGR3
|
anterior gradient 3, protein disulphide isomerase family member
|
|
chr9_-_131270493
Show fit
|
0.71 |
ENST00000372269.7
ENST00000464831.1
|
FAM78A
|
family with sequence similarity 78 member A
|
|
chr2_-_229923163
Show fit
|
0.69 |
ENST00000435716.5
|
TRIP12
|
thyroid hormone receptor interactor 12
|
|
chr4_-_75673360
Show fit
|
0.69 |
ENST00000676584.1
ENST00000678329.1
ENST00000677889.1
ENST00000357854.7
ENST00000359707.9
ENST00000676974.1
ENST00000678273.1
ENST00000677201.1
ENST00000678244.1
ENST00000677125.1
ENST00000677162.1
|
G3BP2
|
G3BP stress granule assembly factor 2
|
|
chr12_+_51912329
Show fit
|
0.68 |
ENST00000547400.5
ENST00000550683.5
ENST00000419526.6
|
ACVRL1
|
activin A receptor like type 1
|
|
chr11_+_33258304
Show fit
|
0.68 |
ENST00000531504.5
ENST00000456517.2
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr12_+_53050014
Show fit
|
0.68 |
ENST00000314250.11
|
TNS2
|
tensin 2
|
|
chr15_+_98648502
Show fit
|
0.68 |
ENST00000650285.1
ENST00000649865.1
|
IGF1R
|
insulin like growth factor 1 receptor
|
|
chr4_+_20251896
Show fit
|
0.67 |
ENST00000504154.6
|
SLIT2
|
slit guidance ligand 2
|
|
chr15_+_70936487
Show fit
|
0.66 |
ENST00000558456.5
ENST00000560158.6
ENST00000558808.5
ENST00000559806.5
ENST00000559069.1
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr12_-_86838867
Show fit
|
0.66 |
ENST00000621808.4
|
MGAT4C
|
MGAT4 family member C
|
|
chr17_+_35121609
Show fit
|
0.65 |
ENST00000158009.6
|
FNDC8
|
fibronectin type III domain containing 8
|
|
chr1_-_182391363
Show fit
|
0.63 |
ENST00000417584.6
|
GLUL
|
glutamate-ammonia ligase
|
|
chr16_+_31074390
Show fit
|
0.62 |
ENST00000300850.5
ENST00000564189.1
ENST00000428260.1
|
ZNF646
|
zinc finger protein 646
|
|
chrX_+_108044967
Show fit
|
0.62 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr11_-_73141275
Show fit
|
0.62 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chr19_-_19515542
Show fit
|
0.61 |
ENST00000585580.4
|
TSSK6
|
testis specific serine kinase 6
|
|
chr4_-_104494882
Show fit
|
0.61 |
ENST00000394767.3
|
CXXC4
|
CXXC finger protein 4
|
|
chr5_-_159099745
Show fit
|
0.60 |
ENST00000517373.1
|
EBF1
|
EBF transcription factor 1
|
|
chr5_-_159099684
Show fit
|
0.60 |
ENST00000380654.8
|
EBF1
|
EBF transcription factor 1
|
|
chr17_+_1762814
Show fit
|
0.60 |
ENST00000570731.5
|
SERPINF1
|
serpin family F member 1
|
|
chrY_+_12904102
Show fit
|
0.59 |
ENST00000360160.8
ENST00000454054.5
|
DDX3Y
|
DEAD-box helicase 3 Y-linked
|
|
chr7_-_131556602
Show fit
|
0.59 |
ENST00000322985.9
ENST00000378555.8
|
PODXL
|
podocalyxin like
|
|
chr9_-_137302264
Show fit
|
0.59 |
ENST00000356628.4
|
NRARP
|
NOTCH regulated ankyrin repeat protein
|
|
chr20_-_3781440
Show fit
|
0.58 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1
|
|
chrX_+_100584928
Show fit
|
0.58 |
ENST00000373031.5
|
TNMD
|
tenomodulin
|
|
chr10_-_60572599
Show fit
|
0.58 |
ENST00000503366.5
|
ANK3
|
ankyrin 3
|
|
chr5_+_139341826
Show fit
|
0.58 |
ENST00000265192.9
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr18_-_55321640
Show fit
|
0.57 |
ENST00000637169.2
|
TCF4
|
transcription factor 4
|
|
chr11_-_46826842
Show fit
|
0.57 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5
|
|
chr5_+_139341875
Show fit
|
0.56 |
ENST00000511706.5
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr17_-_51046868
Show fit
|
0.56 |
ENST00000510283.5
ENST00000510855.1
|
SPAG9
|
sperm associated antigen 9
|
|
chrX_+_108045050
Show fit
|
0.55 |
ENST00000458383.1
ENST00000217957.10
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr7_+_134891566
Show fit
|
0.55 |
ENST00000424922.5
ENST00000495522.1
|
CALD1
|
caldesmon 1
|
|
chr1_+_33864071
Show fit
|
0.55 |
ENST00000681531.1
|
HMGB4
|
high mobility group box 4
|
|
chr14_-_55411817
Show fit
|
0.55 |
ENST00000247178.6
|
ATG14
|
autophagy related 14
|
|
chr2_+_69013282
Show fit
|
0.55 |
ENST00000409829.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1
|
|
chr11_+_112961480
Show fit
|
0.54 |
ENST00000621850.4
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr2_+_86441341
Show fit
|
0.54 |
ENST00000312912.10
ENST00000409064.5
|
KDM3A
|
lysine demethylase 3A
|
|
chr12_+_27245651
Show fit
|
0.54 |
ENST00000543246.5
|
STK38L
|
serine/threonine kinase 38 like
|
|
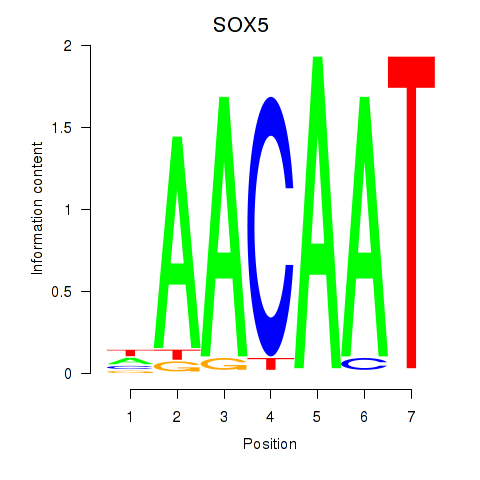
chr12_-_24562438
Show fit
|
0.54 |
ENST00000646273.1
ENST00000659413.1
ENST00000446891.7
|
SOX5
|
SRY-box transcription factor 5
|
|
chr7_+_129375643
Show fit
|
0.54 |
ENST00000490911.5
|
AHCYL2
|
adenosylhomocysteinase like 2
|
|
chr9_-_14314132
Show fit
|
0.53 |
ENST00000380953.6
|
NFIB
|
nuclear factor I B
|
|
chr2_-_110473041
Show fit
|
0.53 |
ENST00000632897.1
ENST00000413601.3
|
LIMS4
|
LIM zinc finger domain containing 4
|
|
chr6_-_56851888
Show fit
|
0.53 |
ENST00000312431.10
ENST00000520645.5
|
DST
|
dystonin
|
|
chr3_+_134795248
Show fit
|
0.52 |
ENST00000398015.8
|
EPHB1
|
EPH receptor B1
|
|
chr7_-_16465728
Show fit
|
0.52 |
ENST00000307068.5
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr7_+_107169720
Show fit
|
0.52 |
ENST00000468401.1
ENST00000497535.5
ENST00000485846.5
|
HBP1
|
HMG-box transcription factor 1
|
|
chr1_-_23014024
Show fit
|
0.51 |
ENST00000440767.2
ENST00000622840.1
|
TEX46
|
testis expressed 46
|
|
chr7_-_140455911
Show fit
|
0.51 |
ENST00000463142.1
|
MKRN1
|
makorin ring finger protein 1
|
|
chr15_+_74619089
Show fit
|
0.51 |
ENST00000562670.5
ENST00000564096.1
|
CLK3
|
CDC like kinase 3
|
|
chr19_+_7516081
Show fit
|
0.51 |
ENST00000597229.2
|
ZNF358
|
zinc finger protein 358
|
|
chr7_-_84195136
Show fit
|
0.50 |
ENST00000420047.1
|
SEMA3A
|
semaphorin 3A
|
|
chr1_+_200739542
Show fit
|
0.50 |
ENST00000358823.6
|
CAMSAP2
|
calmodulin regulated spectrin associated protein family member 2
|
|
chr15_+_83447328
Show fit
|
0.50 |
ENST00000427482.7
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3
|
|
chr8_+_26578399
Show fit
|
0.50 |
ENST00000523027.1
|
DPYSL2
|
dihydropyrimidinase like 2
|
|
chr2_-_55050442
Show fit
|
0.49 |
ENST00000337526.11
|
RTN4
|
reticulon 4
|
|
chr5_-_38595396
Show fit
|
0.49 |
ENST00000263409.8
|
LIFR
|
LIF receptor subunit alpha
|
|
chr13_-_21061492
Show fit
|
0.48 |
ENST00000382592.5
|
LATS2
|
large tumor suppressor kinase 2
|
|
chr15_-_40874216
Show fit
|
0.48 |
ENST00000220507.5
|
RHOV
|
ras homolog family member V
|
|
chr11_-_115504389
Show fit
|
0.48 |
ENST00000545380.5
ENST00000452722.7
ENST00000331581.11
ENST00000537058.5
ENST00000536727.5
ENST00000542447.6
|
CADM1
|
cell adhesion molecule 1
|
|
chr2_-_55050556
Show fit
|
0.47 |
ENST00000394611.6
|
RTN4
|
reticulon 4
|
|
chr4_-_52656536
Show fit
|
0.47 |
ENST00000508499.5
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr7_+_129375830
Show fit
|
0.47 |
ENST00000466993.5
|
AHCYL2
|
adenosylhomocysteinase like 2
|
|
chr5_-_88785493
Show fit
|
0.46 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr11_+_73648674
Show fit
|
0.46 |
ENST00000535129.5
|
PLEKHB1
|
pleckstrin homology domain containing B1
|
|
chr1_-_119811458
Show fit
|
0.46 |
ENST00000256585.10
ENST00000354219.5
ENST00000369401.4
|
REG4
|
regenerating family member 4
|
|
chr17_-_43833137
Show fit
|
0.44 |
ENST00000398389.9
|
MPP3
|
membrane palmitoylated protein 3
|
|
chr3_+_113211539
Show fit
|
0.44 |
ENST00000682979.1
ENST00000485230.5
|
BOC
|
BOC cell adhesion associated, oncogene regulated
|
|
chr2_-_86195400
Show fit
|
0.44 |
ENST00000442664.6
ENST00000409051.6
ENST00000410111.8
ENST00000620815.4
ENST00000449247.6
|
IMMT
|
inner membrane mitochondrial protein
|
|
chr6_+_112236806
Show fit
|
0.44 |
ENST00000588837.5
ENST00000590293.5
ENST00000585450.5
ENST00000629766.2
ENST00000590804.5
ENST00000590584.4
ENST00000628122.2
ENST00000627025.1
ENST00000590673.5
ENST00000585611.5
ENST00000587816.2
|
LAMA4-AS1
ENSG00000281613.2
|
LAMA4 antisense RNA 1
novel ret finger protein-like 4B
|
|
chr4_-_75672868
Show fit
|
0.43 |
ENST00000678123.1
ENST00000678578.1
ENST00000677876.1
ENST00000676839.1
ENST00000678265.1
|
G3BP2
|
G3BP stress granule assembly factor 2
|
|
chr3_+_113211459
Show fit
|
0.43 |
ENST00000495514.5
|
BOC
|
BOC cell adhesion associated, oncogene regulated
|
|
chr3_-_192917832
Show fit
|
0.42 |
ENST00000392452.3
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr9_+_100473140
Show fit
|
0.42 |
ENST00000374879.5
|
TMEFF1
|
transmembrane protein with EGF like and two follistatin like domains 1
|
|
chr4_-_75673139
Show fit
|
0.41 |
ENST00000677566.1
ENST00000503660.5
ENST00000677060.1
ENST00000678552.1
|
G3BP2
|
G3BP stress granule assembly factor 2
|
|
chr3_-_11643871
Show fit
|
0.41 |
ENST00000430365.7
|
VGLL4
|
vestigial like family member 4
|
|
chr22_-_40463411
Show fit
|
0.41 |
ENST00000402630.5
ENST00000407029.7
|
MRTFA
|
myocardin related transcription factor A
|
|
chr15_-_34318761
Show fit
|
0.41 |
ENST00000290209.9
|
SLC12A6
|
solute carrier family 12 member 6
|
|
chrX_-_50814302
Show fit
|
0.41 |
ENST00000289292.11
|
SHROOM4
|
shroom family member 4
|
|
chr14_+_61187544
Show fit
|
0.41 |
ENST00000555185.5
ENST00000557294.5
ENST00000556778.5
|
PRKCH
|
protein kinase C eta
|
|
chr3_-_142028617
Show fit
|
0.40 |
ENST00000477292.5
ENST00000478006.5
ENST00000495310.5
ENST00000486111.5
|
TFDP2
|
transcription factor Dp-2
|
|
chr10_+_35127295
Show fit
|
0.40 |
ENST00000489321.5
ENST00000427847.6
ENST00000374728.7
ENST00000345491.7
ENST00000487132.5
|
CREM
|
cAMP responsive element modulator
|
|
chrX_-_107716401
Show fit
|
0.40 |
ENST00000486554.1
ENST00000372390.8
|
TSC22D3
|
TSC22 domain family member 3
|
|
chr4_-_185811738
Show fit
|
0.40 |
ENST00000451958.5
ENST00000439914.5
ENST00000428330.5
ENST00000429056.5
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr18_+_21363593
Show fit
|
0.39 |
ENST00000580732.6
|
GREB1L
|
GREB1 like retinoic acid receptor coactivator
|
|
chr14_-_104604741
Show fit
|
0.39 |
ENST00000615704.1
ENST00000415614.6
ENST00000556573.6
|
TMEM179
|
transmembrane protein 179
|
|
chr6_+_155216637
Show fit
|
0.38 |
ENST00000275246.11
|
TIAM2
|
TIAM Rac1 associated GEF 2
|
|
chr7_+_48924559
Show fit
|
0.38 |
ENST00000650262.1
|
CDC14C
|
cell division cycle 14C
|
|
chr3_-_142028597
Show fit
|
0.37 |
ENST00000467667.5
|
TFDP2
|
transcription factor Dp-2
|
|
chr14_-_102509798
Show fit
|
0.37 |
ENST00000560748.5
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr12_-_48570046
Show fit
|
0.36 |
ENST00000301046.6
ENST00000549817.1
|
LALBA
|
lactalbumin alpha
|
|
chr1_-_243255320
Show fit
|
0.36 |
ENST00000366544.5
ENST00000366543.5
|
CEP170
|
centrosomal protein 170
|
|
chr13_-_103066411
Show fit
|
0.36 |
ENST00000245312.5
|
SLC10A2
|
solute carrier family 10 member 2
|
|
chr6_+_57090143
Show fit
|
0.36 |
ENST00000508603.5
ENST00000370706.9
ENST00000491832.6
ENST00000370710.10
|
ZNF451
|
zinc finger protein 451
|
|
chr11_+_110130992
Show fit
|
0.35 |
ENST00000528673.5
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr9_-_15510991
Show fit
|
0.35 |
ENST00000380715.5
ENST00000380716.8
ENST00000380738.8
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr1_+_61081728
Show fit
|
0.35 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A
|
|
chr22_+_37696982
Show fit
|
0.34 |
ENST00000644935.1
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr6_+_57090010
Show fit
|
0.34 |
ENST00000510483.5
ENST00000357489.7
|
ZNF451
|
zinc finger protein 451
|
|
chr3_+_134795277
Show fit
|
0.34 |
ENST00000647596.1
|
EPHB1
|
EPH receptor B1
|
|
chr15_+_83447411
Show fit
|
0.34 |
ENST00000324537.5
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3
|
|
chr2_+_109898685
Show fit
|
0.34 |
ENST00000480744.2
|
LIMS3
|
LIM zinc finger domain containing 3
|
|
chr12_-_51083582
Show fit
|
0.34 |
ENST00000548206.1
ENST00000546935.5
ENST00000228515.6
ENST00000548981.5
|
CSRNP2
|
cysteine and serine rich nuclear protein 2
|