Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
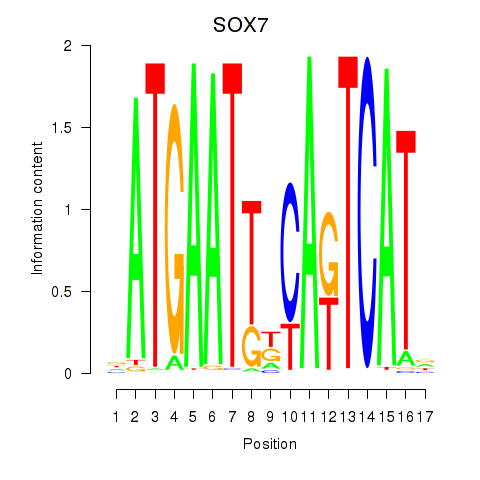
Results for SOX7
Z-value: 0.81
Transcription factors associated with SOX7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX7
|
ENSG00000171056.8 | SRY-box transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX7 | hg38_v1_chr8_-_10730498_10730525 | -0.49 | 4.8e-03 | Click! |
| PINX1 | hg38_v1_chr8_-_10839818_10839855 | -0.43 | 1.4e-02 | Click! |
Activity profile of SOX7 motif
Sorted Z-values of SOX7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_26067622 | 2.70 |
ENST00000374272.4
|
TRIM63
|
tripartite motif containing 63 |
| chr4_-_176269213 | 1.94 |
ENST00000296525.7
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr1_-_89272775 | 1.63 |
ENST00000443807.1
|
GBP5
|
guanylate binding protein 5 |
| chr8_+_28338640 | 1.59 |
ENST00000522209.1
|
PNOC
|
prepronociceptin |
| chr16_+_28292485 | 1.58 |
ENST00000341901.5
|
SBK1
|
SH3 domain binding kinase 1 |
| chr14_-_106117159 | 1.42 |
ENST00000390601.3
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 |
| chr1_+_158931539 | 1.41 |
ENST00000368140.6
ENST00000368138.7 ENST00000392254.6 ENST00000392252.7 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family member 1 |
| chr16_-_20352707 | 1.34 |
ENST00000396134.6
ENST00000573567.5 ENST00000570757.5 ENST00000396138.9 ENST00000571174.5 ENST00000576688.2 |
UMOD
|
uromodulin |
| chr16_-_20352857 | 1.30 |
ENST00000577168.2
|
UMOD
|
uromodulin |
| chr2_-_89222461 | 1.20 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr22_+_22720615 | 1.15 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr16_+_33009175 | 1.14 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr6_+_150368997 | 1.12 |
ENST00000392255.7
ENST00000500320.7 ENST00000344419.8 |
IYD
|
iodotyrosine deiodinase |
| chr17_-_35880350 | 1.06 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr2_-_89100352 | 1.03 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr6_+_150368892 | 1.02 |
ENST00000229447.9
ENST00000392256.6 |
IYD
|
iodotyrosine deiodinase |
| chr16_-_33845229 | 0.99 |
ENST00000569103.2
|
IGHV3OR16-17
|
immunoglobulin heavy variable 3/OR16-17 (non-functional) |
| chr19_-_38426195 | 0.96 |
ENST00000615439.5
ENST00000614135.4 ENST00000622174.4 ENST00000587753.5 ENST00000454404.6 ENST00000617966.4 ENST00000618320.4 ENST00000293062.13 ENST00000433821.6 ENST00000426920.6 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr4_+_76074701 | 0.84 |
ENST00000355810.9
ENST00000349321.7 |
ART3
|
ADP-ribosyltransferase 3 (inactive) |
| chr15_+_63122561 | 0.79 |
ENST00000557972.1
|
LACTB
|
lactamase beta |
| chr6_+_42563981 | 0.79 |
ENST00000372899.6
ENST00000372901.2 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr2_+_86907953 | 0.78 |
ENST00000409776.6
|
RGPD1
|
RANBP2 like and GRIP domain containing 1 |
| chr12_-_56299974 | 0.70 |
ENST00000547298.5
ENST00000551936.5 ENST00000551253.5 ENST00000551473.5 |
CS
|
citrate synthase |
| chr4_-_122456725 | 0.68 |
ENST00000226730.5
|
IL2
|
interleukin 2 |
| chr6_+_143843316 | 0.66 |
ENST00000367576.6
|
LTV1
|
LTV1 ribosome biogenesis factor |
| chr12_-_89656093 | 0.66 |
ENST00000359142.7
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr6_+_116399395 | 0.64 |
ENST00000644499.1
|
ENSG00000285446.1
|
novel protein |
| chr13_-_46020487 | 0.64 |
ENST00000464597.2
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr19_-_38426162 | 0.63 |
ENST00000587738.2
ENST00000586305.5 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr12_-_89656051 | 0.61 |
ENST00000261173.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr12_-_10420550 | 0.56 |
ENST00000381903.2
ENST00000396439.7 |
KLRC3
|
killer cell lectin like receptor C3 |
| chr13_-_37059432 | 0.54 |
ENST00000464744.5
|
SUPT20H
|
SPT20 homolog, SAGA complex component |
| chr5_+_162067858 | 0.51 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chrX_+_131083706 | 0.51 |
ENST00000370921.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr3_-_71306012 | 0.47 |
ENST00000649431.1
ENST00000610810.5 |
FOXP1
|
forkhead box P1 |
| chr4_+_146214515 | 0.46 |
ENST00000636502.1
|
REELD1
|
reeler domain containing 1 |
| chr14_+_60245744 | 0.45 |
ENST00000325642.7
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent 1A |
| chr5_+_162067764 | 0.44 |
ENST00000639213.2
ENST00000414552.6 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr11_+_35186820 | 0.43 |
ENST00000531110.6
ENST00000525685.6 |
CD44
|
CD44 molecule (Indian blood group) |
| chr3_-_71305986 | 0.42 |
ENST00000647614.1
|
FOXP1
|
forkhead box P1 |
| chr5_+_162067500 | 0.40 |
ENST00000639384.1
ENST00000640985.1 ENST00000638772.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr11_+_5689691 | 0.39 |
ENST00000425490.5
|
TRIM22
|
tripartite motif containing 22 |
| chr1_-_186461089 | 0.38 |
ENST00000391997.3
|
PDC
|
phosducin |
| chr1_-_158426237 | 0.37 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr5_+_162067458 | 0.31 |
ENST00000639975.1
ENST00000639111.2 ENST00000639683.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr6_+_29100609 | 0.29 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor family 2 subfamily J member 1 |
| chr19_+_37346316 | 0.27 |
ENST00000591134.5
|
ZNF875
|
zinc finger protein 875 |
| chr17_+_82900542 | 0.27 |
ENST00000574422.1
|
TBCD
|
tubulin folding cofactor D |
| chr7_+_134565098 | 0.26 |
ENST00000652743.1
|
AKR1B15
|
aldo-keto reductase family 1 member B15 |
| chr10_+_95911254 | 0.24 |
ENST00000636965.1
|
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr11_-_6003194 | 0.24 |
ENST00000330728.4
ENST00000641279.1 |
OR56A4
|
olfactory receptor family 56 subfamily A member 4 |
| chr17_-_30334050 | 0.23 |
ENST00000328886.5
ENST00000538566.6 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr12_+_55492378 | 0.23 |
ENST00000548615.1
|
OR6C68
|
olfactory receptor family 6 subfamily C member 68 |
| chr7_+_141918817 | 0.23 |
ENST00000548136.1
|
OR9A4
|
olfactory receptor family 9 subfamily A member 4 |
| chr11_+_5754243 | 0.23 |
ENST00000641350.2
|
OR52N4
|
olfactory receptor family 52 subfamily N member 4 |
| chr17_-_67239800 | 0.22 |
ENST00000579861.1
|
HELZ
|
helicase with zinc finger |
| chr11_-_62707331 | 0.22 |
ENST00000533982.1
|
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chr3_+_98463201 | 0.19 |
ENST00000642057.1
|
OR5K1
|
olfactory receptor family 5 subfamily K member 1 |
| chr14_-_23104845 | 0.19 |
ENST00000642668.1
ENST00000644000.1 |
LMLN2
|
leishmanolysin like peptidase 2 |
| chr11_+_7597182 | 0.19 |
ENST00000528883.5
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr12_-_56934403 | 0.16 |
ENST00000293502.2
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C member 7 |
| chr18_+_35241027 | 0.16 |
ENST00000330501.12
ENST00000601719.1 ENST00000591206.5 ENST00000261333.10 ENST00000355632.8 ENST00000585800.1 |
ZNF397
|
zinc finger protein 397 |
| chr4_+_74308492 | 0.15 |
ENST00000502358.5
ENST00000503098.5 ENST00000505212.5 ENST00000509145.5 ENST00000514968.5 |
EPGN
|
epithelial mitogen |
| chr12_-_10998304 | 0.15 |
ENST00000538986.2
|
TAS2R20
|
taste 2 receptor member 20 |
| chr8_-_19602484 | 0.15 |
ENST00000454498.6
ENST00000520003.5 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr6_-_111483700 | 0.15 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr1_-_152089062 | 0.15 |
ENST00000368806.2
|
TCHHL1
|
trichohyalin like 1 |
| chr19_+_34926892 | 0.14 |
ENST00000303586.11
ENST00000601142.2 ENST00000439785.5 ENST00000601540.5 ENST00000601957.5 |
ZNF30
|
zinc finger protein 30 |
| chr8_+_12945667 | 0.14 |
ENST00000524591.7
|
TRMT9B
|
tRNA methyltransferase 9B (putative) |
| chr19_-_5784599 | 0.13 |
ENST00000390672.2
ENST00000419421.3 |
PRR22
|
proline rich 22 |
| chr1_+_247738615 | 0.12 |
ENST00000283225.2
|
OR14K1
|
olfactory receptor family 14 subfamily K member 1 |
| chr21_-_14546297 | 0.11 |
ENST00000400566.6
ENST00000400564.5 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr1_+_200027731 | 0.10 |
ENST00000447034.1
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chr3_-_122564577 | 0.10 |
ENST00000477522.6
ENST00000360356.6 |
PARP9
|
poly(ADP-ribose) polymerase family member 9 |
| chr12_-_123243998 | 0.09 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr1_+_100133186 | 0.06 |
ENST00000370139.1
|
TRMT13
|
tRNA methyltransferase 13 homolog |
| chr10_+_24449426 | 0.06 |
ENST00000307544.10
|
KIAA1217
|
KIAA1217 |
| chr3_-_36993103 | 0.06 |
ENST00000322716.8
|
EPM2AIP1
|
EPM2A interacting protein 1 |
| chr7_-_138755892 | 0.05 |
ENST00000644341.1
ENST00000478480.2 |
ATP6V0A4
|
ATPase H+ transporting V0 subunit a4 |
| chr2_-_105396750 | 0.05 |
ENST00000447958.1
|
FHL2
|
four and a half LIM domains 2 |
| chr7_+_100120026 | 0.04 |
ENST00000483756.5
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr3_-_161371501 | 0.03 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase small subunit B |
| chr4_-_47981535 | 0.03 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr13_+_94601830 | 0.02 |
ENST00000376958.5
|
GPR180
|
G protein-coupled receptor 180 |
| chr8_-_86069662 | 0.02 |
ENST00000276616.3
|
PSKH2
|
protein serine kinase H2 |
| chr10_-_12042771 | 0.01 |
ENST00000357604.10
|
UPF2
|
UPF2 regulator of nonsense mediated mRNA decay |
| chr4_+_74308487 | 0.00 |
ENST00000332112.8
|
EPGN
|
epithelial mitogen |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.6 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.5 | 2.7 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.3 | 1.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 1.6 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.2 | 0.8 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 2.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.1 | 1.3 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 0.7 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.4 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 1.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.9 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 5.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.6 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.4 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.8 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 1.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 2.9 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 2.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 1.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.7 | GO:0060170 | ciliary membrane(GO:0060170) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.2 | 0.7 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.2 | 1.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 1.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 2.6 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 2.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 1.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.7 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 1.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 5.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 5.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |