Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
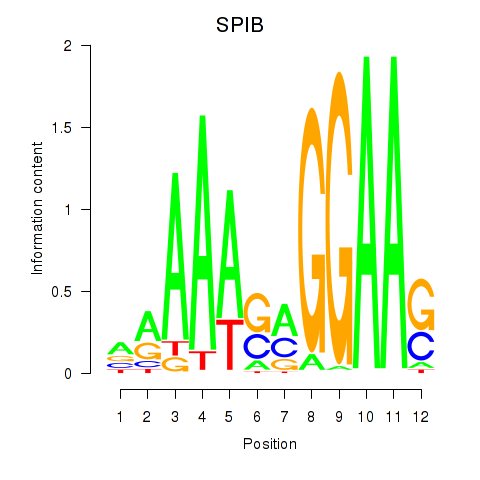
Results for SPIB
Z-value: 5.65
Transcription factors associated with SPIB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIB
|
ENSG00000269404.7 | Spi-B transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPIB | hg38_v1_chr19_+_50418930_50418958 | 0.74 | 1.5e-06 | Click! |
Activity profile of SPIB motif
Sorted Z-values of SPIB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_160711803 | 44.00 |
ENST00000368045.3
ENST00000368046.8 ENST00000613788.1 |
CD48
|
CD48 molecule |
| chr17_-_74712911 | 35.38 |
ENST00000326165.11
ENST00000583937.5 ENST00000301573.13 ENST00000469092.5 |
CD300LF
|
CD300 molecule like family member f |
| chr22_+_36913620 | 30.72 |
ENST00000403662.8
ENST00000262825.9 |
CSF2RB
|
colony stimulating factor 2 receptor subunit beta |
| chrX_-_47629845 | 28.61 |
ENST00000469388.1
ENST00000396992.8 ENST00000377005.6 |
CFP
|
complement factor properdin |
| chr19_+_41877267 | 27.50 |
ENST00000221972.8
ENST00000597454.1 ENST00000444740.2 |
CD79A
|
CD79a molecule |
| chr17_-_5234801 | 26.93 |
ENST00000571800.5
ENST00000574081.6 ENST00000399600.8 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr7_+_74773962 | 25.59 |
ENST00000289473.10
|
NCF1
|
neutrophil cytosolic factor 1 |
| chr16_+_30183595 | 25.01 |
ENST00000219150.10
ENST00000570045.5 ENST00000565497.5 ENST00000570244.5 |
CORO1A
|
coronin 1A |
| chr17_-_74623730 | 23.85 |
ENST00000392619.2
|
CD300E
|
CD300e molecule |
| chr3_-_151329539 | 23.36 |
ENST00000325602.6
|
P2RY13
|
purinergic receptor P2Y13 |
| chr5_+_169637241 | 23.03 |
ENST00000520908.7
|
DOCK2
|
dedicator of cytokinesis 2 |
| chrX_+_78945332 | 22.98 |
ENST00000544091.1
ENST00000171757.3 |
P2RY10
|
P2Y receptor family member 10 |
| chr12_+_9971402 | 22.60 |
ENST00000304361.9
ENST00000396507.7 ENST00000434319.6 |
CLEC12A
|
C-type lectin domain family 12 member A |
| chr7_-_36724457 | 22.14 |
ENST00000617537.5
ENST00000435386.1 |
AOAH
|
acyloxyacyl hydrolase |
| chr1_+_161706949 | 22.02 |
ENST00000350710.3
ENST00000367949.6 ENST00000367959.6 ENST00000540521.5 ENST00000546024.5 ENST00000674251.1 ENST00000674323.1 |
FCRLA
|
Fc receptor like A |
| chr12_+_8123837 | 21.46 |
ENST00000345999.9
ENST00000352620.9 ENST00000360500.5 |
CLEC4A
|
C-type lectin domain family 4 member A |
| chr17_-_31314040 | 21.45 |
ENST00000330927.5
|
EVI2B
|
ecotropic viral integration site 2B |
| chr11_-_67437670 | 21.34 |
ENST00000326294.4
|
PTPRCAP
|
protein tyrosine phosphatase receptor type C associated protein |
| chr1_+_111473792 | 21.30 |
ENST00000343534.9
|
C1orf162
|
chromosome 1 open reading frame 162 |
| chr1_-_30757767 | 21.20 |
ENST00000294507.4
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr19_+_16143678 | 21.12 |
ENST00000613986.4
ENST00000593031.1 |
HSH2D
|
hematopoietic SH2 domain containing |
| chr6_-_24935942 | 21.06 |
ENST00000645100.1
ENST00000643898.2 ENST00000613507.4 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr17_-_31314066 | 20.81 |
ENST00000577894.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr1_+_209768482 | 20.69 |
ENST00000367023.5
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr5_+_55102635 | 20.27 |
ENST00000274306.7
|
GZMA
|
granzyme A |
| chr7_-_36724380 | 20.11 |
ENST00000617267.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr1_+_111473972 | 19.67 |
ENST00000369718.4
|
C1orf162
|
chromosome 1 open reading frame 162 |
| chr2_+_68365274 | 19.65 |
ENST00000234313.8
|
PLEK
|
pleckstrin |
| chr7_-_36724543 | 19.57 |
ENST00000612871.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr6_-_159045104 | 18.85 |
ENST00000326965.7
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr1_+_26317950 | 18.77 |
ENST00000374213.3
|
CD52
|
CD52 molecule |
| chr6_-_159044980 | 18.76 |
ENST00000367066.8
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr12_-_14961256 | 18.69 |
ENST00000541380.5
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr6_-_132757883 | 18.24 |
ENST00000525289.5
ENST00000326499.11 |
VNN2
|
vanin 2 |
| chr1_+_161505412 | 18.01 |
ENST00000367972.8
|
FCGR2A
|
Fc fragment of IgG receptor IIa |
| chr5_+_35856883 | 17.92 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr12_+_9971512 | 17.81 |
ENST00000350667.4
|
CLEC12A
|
C-type lectin domain family 12 member A |
| chr16_-_50681289 | 17.68 |
ENST00000423026.6
ENST00000330943.9 |
SNX20
|
sorting nexin 20 |
| chr6_+_391743 | 17.59 |
ENST00000380956.9
|
IRF4
|
interferon regulatory factor 4 |
| chr7_+_50304038 | 17.39 |
ENST00000642219.1
ENST00000645066.1 |
IKZF1
|
IKAROS family zinc finger 1 |
| chr17_-_8965674 | 17.36 |
ENST00000447110.6
ENST00000611902.4 ENST00000616147.4 ENST00000623421.3 |
PIK3R5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr6_-_159045010 | 17.33 |
ENST00000338313.5
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr7_+_139829242 | 17.13 |
ENST00000455353.6
ENST00000458722.6 ENST00000448866.7 ENST00000411653.6 |
TBXAS1
|
thromboxane A synthase 1 |
| chr19_-_54222978 | 16.97 |
ENST00000245620.13
ENST00000346401.10 ENST00000445347.1 |
LILRB3
|
leukocyte immunoglobulin like receptor B3 |
| chr9_-_35618367 | 16.88 |
ENST00000378431.5
ENST00000378430.3 ENST00000259633.9 |
CD72
|
CD72 molecule |
| chr20_+_59163810 | 16.83 |
ENST00000371030.4
|
ZNF831
|
zinc finger protein 831 |
| chr19_+_6772699 | 16.82 |
ENST00000602142.6
ENST00000304076.6 ENST00000596764.5 |
VAV1
|
vav guanine nucleotide exchange factor 1 |
| chr3_+_122055355 | 16.73 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr6_-_41286665 | 16.33 |
ENST00000589614.5
ENST00000244709.9 ENST00000334475.10 ENST00000591620.1 |
TREM1
|
triggering receptor expressed on myeloid cells 1 |
| chr11_-_59212869 | 16.33 |
ENST00000361050.4
|
MPEG1
|
macrophage expressed 1 |
| chr20_-_64079479 | 16.26 |
ENST00000395042.2
|
RGS19
|
regulator of G protein signaling 19 |
| chr22_-_37244237 | 16.24 |
ENST00000401529.3
ENST00000249071.11 |
RAC2
|
Rac family small GTPase 2 |
| chr20_-_64079906 | 16.19 |
ENST00000332298.9
|
RGS19
|
regulator of G protein signaling 19 |
| chr11_-_64745331 | 16.18 |
ENST00000377489.5
ENST00000354024.7 |
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr12_-_54984667 | 16.09 |
ENST00000524668.5
ENST00000533607.1 ENST00000449076.6 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr15_+_76995118 | 15.82 |
ENST00000558012.6
ENST00000379595.7 |
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr11_-_64744811 | 15.57 |
ENST00000377497.7
ENST00000377487.5 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr7_+_139829153 | 15.52 |
ENST00000652056.1
|
TBXAS1
|
thromboxane A synthase 1 |
| chr7_+_150514851 | 15.48 |
ENST00000313543.5
|
GIMAP7
|
GTPase, IMAP family member 7 |
| chr1_+_161707244 | 15.21 |
ENST00000349527.8
ENST00000294796.8 ENST00000309691.10 ENST00000367953.7 ENST00000367950.2 |
FCRLA
|
Fc receptor like A |
| chr16_+_3046552 | 15.16 |
ENST00000336577.9
|
MMP25
|
matrix metallopeptidase 25 |
| chr13_+_108269629 | 15.05 |
ENST00000430559.5
ENST00000375887.9 |
TNFSF13B
|
TNF superfamily member 13b |
| chr14_+_88005128 | 14.97 |
ENST00000267549.5
|
GPR65
|
G protein-coupled receptor 65 |
| chr9_+_90801757 | 14.86 |
ENST00000375751.8
ENST00000375754.9 |
SYK
|
spleen associated tyrosine kinase |
| chr12_+_54497712 | 14.86 |
ENST00000293373.11
|
NCKAP1L
|
NCK associated protein 1 like |
| chrX_-_30577759 | 14.85 |
ENST00000378962.4
|
TASL
|
TLR adaptor interacting with endolysosomal SLC15A4 |
| chr5_+_58491427 | 14.79 |
ENST00000396776.6
ENST00000502276.6 ENST00000511930.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr12_-_14961610 | 14.72 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr19_+_48325522 | 14.65 |
ENST00000594198.1
ENST00000270221.11 ENST00000597279.5 ENST00000593437.1 |
EMP3
|
epithelial membrane protein 3 |
| chr1_-_160647037 | 14.64 |
ENST00000302035.11
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr12_-_14961559 | 14.58 |
ENST00000228945.9
ENST00000541546.5 |
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr9_+_214843 | 14.57 |
ENST00000432829.7
|
DOCK8
|
dedicator of cytokinesis 8 |
| chr12_+_6951345 | 14.32 |
ENST00000318974.14
ENST00000541698.5 ENST00000542462.1 |
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chr5_-_67196791 | 13.94 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr10_+_26438317 | 13.80 |
ENST00000376236.9
|
APBB1IP
|
amyloid beta precursor protein binding family B member 1 interacting protein |
| chr3_+_46354072 | 13.76 |
ENST00000445132.3
ENST00000421659.1 |
CCR2
|
C-C motif chemokine receptor 2 |
| chr13_+_30735523 | 13.71 |
ENST00000380490.5
|
ALOX5AP
|
arachidonate 5-lipoxygenase activating protein |
| chr3_+_122077850 | 13.64 |
ENST00000482356.5
ENST00000393627.6 |
CD86
|
CD86 molecule |
| chr22_+_36922040 | 13.39 |
ENST00000406230.5
|
CSF2RB
|
colony stimulating factor 2 receptor subunit beta |
| chr11_-_3837858 | 13.27 |
ENST00000396979.1
|
RHOG
|
ras homolog family member G |
| chr9_+_90801909 | 13.24 |
ENST00000375747.5
|
SYK
|
spleen associated tyrosine kinase |
| chr2_+_143129379 | 13.24 |
ENST00000295095.11
|
ARHGAP15
|
Rho GTPase activating protein 15 |
| chr5_+_58491451 | 13.17 |
ENST00000513924.2
ENST00000515443.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr3_+_32391871 | 13.13 |
ENST00000465248.1
|
CMTM7
|
CKLF like MARVEL transmembrane domain containing 7 |
| chr12_+_6951271 | 13.05 |
ENST00000456013.5
|
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chrX_-_130110679 | 13.01 |
ENST00000335997.11
|
ELF4
|
E74 like ETS transcription factor 4 |
| chr5_+_119333151 | 12.96 |
ENST00000513374.1
|
TNFAIP8
|
TNF alpha induced protein 8 |
| chr19_+_48325323 | 12.96 |
ENST00000596315.5
|
EMP3
|
epithelial membrane protein 3 |
| chr11_-_47378391 | 12.94 |
ENST00000227163.8
|
SPI1
|
Spi-1 proto-oncogene |
| chr17_+_74671096 | 12.75 |
ENST00000402449.8
|
RAB37
|
RAB37, member RAS oncogene family |
| chr11_-_47378527 | 12.63 |
ENST00000378538.8
|
SPI1
|
Spi-1 proto-oncogene |
| chrX_-_78327604 | 12.49 |
ENST00000373304.4
|
CYSLTR1
|
cysteinyl leukotriene receptor 1 |
| chr11_-_47378494 | 12.47 |
ENST00000533030.1
|
SPI1
|
Spi-1 proto-oncogene |
| chr22_-_37244417 | 12.45 |
ENST00000405484.5
ENST00000441619.5 ENST00000406508.5 |
RAC2
|
Rac family small GTPase 2 |
| chrX_-_78327626 | 12.43 |
ENST00000614798.1
|
CYSLTR1
|
cysteinyl leukotriene receptor 1 |
| chr1_+_161581339 | 12.40 |
ENST00000543859.5
ENST00000611236.1 |
FCGR2C
|
Fc fragment of IgG receptor IIc (gene/pseudogene) |
| chr12_-_9733292 | 12.36 |
ENST00000621400.4
ENST00000327839.3 |
CLECL1
|
C-type lectin like 1 |
| chr1_+_161505438 | 12.31 |
ENST00000271450.12
|
FCGR2A
|
Fc fragment of IgG receptor IIa |
| chr6_+_37005630 | 12.31 |
ENST00000274963.13
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr14_-_22819721 | 12.27 |
ENST00000554517.5
ENST00000285850.11 ENST00000397529.6 ENST00000555702.5 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr19_-_10335773 | 12.23 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr3_+_122077776 | 12.20 |
ENST00000264468.9
|
CD86
|
CD86 molecule |
| chr6_+_31586124 | 12.19 |
ENST00000418507.6
ENST00000376096.5 ENST00000376099.5 ENST00000376110.7 |
LST1
|
leukocyte specific transcript 1 |
| chr5_-_39270623 | 12.19 |
ENST00000512138.1
ENST00000646045.2 |
FYB1
|
FYN binding protein 1 |
| chr19_-_38426195 | 12.16 |
ENST00000615439.5
ENST00000614135.4 ENST00000622174.4 ENST00000587753.5 ENST00000454404.6 ENST00000617966.4 ENST00000618320.4 ENST00000293062.13 ENST00000433821.6 ENST00000426920.6 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr1_-_160646958 | 12.08 |
ENST00000538290.2
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr1_+_206470463 | 11.99 |
ENST00000581977.7
ENST00000578328.6 ENST00000584998.5 ENST00000579827.6 |
IKBKE
|
inhibitor of nuclear factor kappa B kinase subunit epsilon |
| chr11_-_47378479 | 11.95 |
ENST00000533968.1
|
SPI1
|
Spi-1 proto-oncogene |
| chr12_+_6951250 | 11.84 |
ENST00000538715.5
|
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chr10_-_96720485 | 11.81 |
ENST00000339364.10
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr17_-_19046957 | 11.81 |
ENST00000284154.10
ENST00000573099.5 |
GRAP
|
GRB2 related adaptor protein |
| chr6_+_31615215 | 11.81 |
ENST00000337917.11
ENST00000376059.8 |
AIF1
|
allograft inflammatory factor 1 |
| chr9_+_215157 | 11.78 |
ENST00000479404.5
|
DOCK8
|
dedicator of cytokinesis 8 |
| chr21_-_44920892 | 11.71 |
ENST00000397846.7
ENST00000652462.1 ENST00000302347.10 ENST00000524251.1 |
ITGB2
|
integrin subunit beta 2 |
| chr6_+_31586680 | 11.71 |
ENST00000339530.8
|
LST1
|
leukocyte specific transcript 1 |
| chr20_-_36646146 | 11.65 |
ENST00000262866.9
|
SLA2
|
Src like adaptor 2 |
| chr6_+_31586835 | 11.59 |
ENST00000211921.11
|
LST1
|
leukocyte specific transcript 1 |
| chrX_-_130110479 | 11.35 |
ENST00000308167.10
|
ELF4
|
E74 like ETS transcription factor 4 |
| chr1_+_151156627 | 11.30 |
ENST00000368910.4
|
TNFAIP8L2
|
TNF alpha induced protein 8 like 2 |
| chr8_+_73991345 | 11.04 |
ENST00000284818.7
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr4_+_67558719 | 11.03 |
ENST00000265404.7
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr19_-_3786254 | 11.02 |
ENST00000585778.5
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr19_+_51225059 | 10.95 |
ENST00000436584.6
ENST00000421133.6 ENST00000262262.5 ENST00000391796.7 |
CD33
|
CD33 molecule |
| chr1_+_149782671 | 10.94 |
ENST00000444948.5
ENST00000369168.5 |
FCGR1A
|
Fc fragment of IgG receptor Ia |
| chr6_+_31586269 | 10.87 |
ENST00000438075.7
|
LST1
|
leukocyte specific transcript 1 |
| chr17_-_75393870 | 10.83 |
ENST00000582582.1
|
GRB2
|
growth factor receptor bound protein 2 |
| chr3_-_121660892 | 10.80 |
ENST00000428394.6
ENST00000314583.8 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr2_+_233059838 | 10.79 |
ENST00000359570.9
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr1_+_159302321 | 10.60 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE receptor Ia |
| chr21_-_44920855 | 10.53 |
ENST00000397854.7
|
ITGB2
|
integrin subunit beta 2 |
| chr1_-_17439657 | 10.51 |
ENST00000375436.9
|
RCC2
|
regulator of chromosome condensation 2 |
| chr17_-_40937445 | 10.49 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr19_-_13102848 | 10.48 |
ENST00000264824.5
|
LYL1
|
LYL1 basic helix-loop-helix family member |
| chr6_+_31587049 | 10.44 |
ENST00000376089.6
ENST00000396112.6 |
LST1
|
leukocyte specific transcript 1 |
| chr13_+_108269880 | 10.34 |
ENST00000542136.1
|
TNFSF13B
|
TNF superfamily member 13b |
| chr13_+_30713477 | 10.20 |
ENST00000617770.4
|
ALOX5AP
|
arachidonate 5-lipoxygenase activating protein |
| chr16_-_50681206 | 10.19 |
ENST00000610485.1
|
SNX20
|
sorting nexin 20 |
| chr6_+_31587002 | 10.10 |
ENST00000376090.6
|
LST1
|
leukocyte specific transcript 1 |
| chr17_-_40937641 | 10.09 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr9_-_127778659 | 9.89 |
ENST00000314830.13
|
SH2D3C
|
SH2 domain containing 3C |
| chr11_+_60378475 | 9.86 |
ENST00000358246.5
|
MS4A7
|
membrane spanning 4-domains A7 |
| chr19_-_3786363 | 9.81 |
ENST00000310132.11
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr11_+_60378494 | 9.81 |
ENST00000534016.5
|
MS4A7
|
membrane spanning 4-domains A7 |
| chr2_+_113127588 | 9.79 |
ENST00000409930.4
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr1_+_161707222 | 9.74 |
ENST00000236938.12
|
FCRLA
|
Fc receptor like A |
| chr15_+_76995374 | 9.72 |
ENST00000559161.5
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr2_+_113173950 | 9.63 |
ENST00000245796.11
ENST00000441564.7 |
PSD4
|
pleckstrin and Sec7 domain containing 4 |
| chr17_-_75393656 | 9.54 |
ENST00000392563.5
|
GRB2
|
growth factor receptor bound protein 2 |
| chr1_+_161707205 | 9.34 |
ENST00000367957.7
|
FCRLA
|
Fc receptor like A |
| chr16_+_11965193 | 9.32 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr11_+_60378524 | 9.31 |
ENST00000530614.5
ENST00000530027.5 ENST00000300184.8 ENST00000530234.2 ENST00000528215.1 ENST00000531787.5 |
MS4A7
MS4A14
|
membrane spanning 4-domains A7 membrane spanning 4-domains A14 |
| chr17_-_75393741 | 9.26 |
ENST00000578961.5
ENST00000392564.5 |
GRB2
|
growth factor receptor bound protein 2 |
| chr21_-_44920918 | 9.16 |
ENST00000522688.5
|
ITGB2
|
integrin subunit beta 2 |
| chr8_-_100722174 | 9.10 |
ENST00000677380.1
ENST00000677478.1 |
PABPC1
|
poly(A) binding protein cytoplasmic 1 |
| chr12_+_25052732 | 9.08 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr19_-_51417619 | 9.05 |
ENST00000441969.7
ENST00000339313.10 ENST00000525998.5 ENST00000436984.6 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr7_+_106865263 | 9.04 |
ENST00000440650.6
ENST00000496166.6 ENST00000473541.5 |
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr19_-_3786408 | 9.04 |
ENST00000395040.6
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr2_+_233060295 | 9.03 |
ENST00000445964.6
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr11_+_64340191 | 9.00 |
ENST00000356786.10
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr16_+_57542672 | 8.99 |
ENST00000615867.4
ENST00000340339.4 |
ADGRG5
|
adhesion G protein-coupled receptor G5 |
| chr1_+_89821921 | 8.97 |
ENST00000394593.7
|
LRRC8D
|
leucine rich repeat containing 8 VRAC subunit D |
| chr19_+_54630410 | 8.87 |
ENST00000396327.7
ENST00000324602.12 |
LILRB1
|
leukocyte immunoglobulin like receptor B1 |
| chr14_+_22007503 | 8.80 |
ENST00000390447.3
|
TRAV19
|
T cell receptor alpha variable 19 |
| chr19_+_35154914 | 8.73 |
ENST00000423817.7
|
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr1_+_117001744 | 8.60 |
ENST00000256652.8
ENST00000682167.1 ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr2_-_144430934 | 8.53 |
ENST00000638087.1
ENST00000638007.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr10_+_26438404 | 8.52 |
ENST00000356785.4
|
APBB1IP
|
amyloid beta precursor protein binding family B member 1 interacting protein |
| chr8_-_100722036 | 8.51 |
ENST00000518196.5
ENST00000519004.5 ENST00000519363.1 ENST00000318607.10 ENST00000520142.2 |
PABPC1
|
poly(A) binding protein cytoplasmic 1 |
| chr2_-_230219944 | 8.49 |
ENST00000455674.2
ENST00000392048.7 ENST00000258381.11 ENST00000358662.9 ENST00000258382.10 |
SP110
|
SP110 nuclear body protein |
| chr2_-_74178802 | 8.26 |
ENST00000396049.5
|
MOB1A
|
MOB kinase activator 1A |
| chr1_-_159923717 | 8.22 |
ENST00000368096.5
|
TAGLN2
|
transgelin 2 |
| chr2_-_144431001 | 8.22 |
ENST00000636413.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr22_-_23838987 | 8.06 |
ENST00000318109.12
ENST00000404056.1 ENST00000406855.7 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr10_+_95755737 | 8.04 |
ENST00000543964.6
|
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr1_+_121087343 | 7.97 |
ENST00000616817.4
ENST00000623603.3 ENST00000369384.9 ENST00000369383.8 ENST00000369178.5 |
FCGR1B
|
Fc fragment of IgG receptor Ib |
| chr7_-_116159886 | 7.85 |
ENST00000484212.5
|
TFEC
|
transcription factor EC |
| chr6_-_30684744 | 7.85 |
ENST00000615892.4
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr5_-_157109025 | 7.84 |
ENST00000307851.9
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr12_-_121802886 | 7.76 |
ENST00000545885.5
ENST00000542933.5 ENST00000428029.6 ENST00000541694.5 ENST00000536662.5 ENST00000535643.5 ENST00000541657.5 |
LINC01089
RHOF
|
long intergenic non-protein coding RNA 1089 ras homolog family member F, filopodia associated |
| chr19_+_10848366 | 7.66 |
ENST00000397820.5
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr17_-_45410414 | 7.65 |
ENST00000532038.5
ENST00000528677.1 |
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr15_-_34337462 | 7.58 |
ENST00000676379.1
|
SLC12A6
|
solute carrier family 12 member 6 |
| chr19_+_54630497 | 7.57 |
ENST00000396332.8
ENST00000427581.6 |
LILRB1
|
leukocyte immunoglobulin like receptor B1 |
| chr1_+_161663147 | 7.56 |
ENST00000236937.13
ENST00000367961.8 ENST00000358671.9 |
FCGR2B
|
Fc fragment of IgG receptor IIb |
| chr6_+_116461364 | 7.47 |
ENST00000368606.7
ENST00000368605.3 |
CALHM6
|
calcium homeostasis modulator family member 6 |
| chr19_-_13103151 | 7.45 |
ENST00000590974.1
|
LYL1
|
LYL1 basic helix-loop-helix family member |
| chr17_+_19127535 | 7.44 |
ENST00000577213.1
ENST00000344415.9 |
GRAPL
|
GRB2 related adaptor protein like |
| chr16_+_57542635 | 7.40 |
ENST00000349457.8
|
ADGRG5
|
adhesion G protein-coupled receptor G5 |
| chr10_-_119542683 | 7.40 |
ENST00000369103.3
|
RGS10
|
regulator of G protein signaling 10 |
| chr5_-_157109115 | 7.36 |
ENST00000522593.5
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr16_+_10866247 | 7.26 |
ENST00000636238.1
|
CIITA
|
class II major histocompatibility complex transactivator |
| chr1_-_153946652 | 7.24 |
ENST00000361217.9
|
DENND4B
|
DENN domain containing 4B |
| chrX_-_130110361 | 7.14 |
ENST00000434609.1
|
ELF4
|
E74 like ETS transcription factor 4 |
| chr19_-_54242751 | 7.13 |
ENST00000245621.6
ENST00000396365.6 |
LILRA6
|
leukocyte immunoglobulin like receptor A6 |
| chr16_-_50681328 | 7.09 |
ENST00000300590.7
|
SNX20
|
sorting nexin 20 |
| chr5_-_140633639 | 7.00 |
ENST00000498971.6
|
CD14
|
CD14 molecule |
| chr5_-_140633690 | 6.97 |
ENST00000512545.1
ENST00000401743.6 |
CD14
|
CD14 molecule |
| chr8_-_100721851 | 6.92 |
ENST00000522658.6
|
PABPC1
|
poly(A) binding protein cytoplasmic 1 |
| chr19_-_51417581 | 6.76 |
ENST00000442846.7
ENST00000530476.1 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIB
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.0 | 60.2 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 13.8 | 13.8 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 11.8 | 35.4 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 11.0 | 11.0 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 10.0 | 50.0 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 9.7 | 38.7 | GO:0045399 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) |
| 8.8 | 26.4 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 8.3 | 49.7 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 8.0 | 48.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 7.1 | 50.0 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 6.5 | 39.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 6.3 | 25.4 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 5.9 | 47.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 5.1 | 15.2 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) |
| 5.0 | 25.0 | GO:0032796 | uropod organization(GO:0032796) |
| 5.0 | 19.8 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 5.0 | 14.9 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 4.4 | 13.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 4.1 | 16.4 | GO:0035548 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 4.1 | 28.6 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 3.8 | 7.6 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 3.5 | 21.1 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 3.4 | 30.7 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 2.9 | 32.4 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 2.9 | 8.7 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 2.8 | 19.6 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 2.8 | 24.9 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 2.7 | 16.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 2.6 | 15.7 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 2.6 | 28.7 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 2.5 | 34.8 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 2.4 | 11.8 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 2.3 | 21.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 2.3 | 16.3 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 2.2 | 17.9 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 2.2 | 22.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 2.1 | 6.4 | GO:1904266 | positive regulation of Schwann cell migration(GO:1900149) regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 2.0 | 4.1 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 1.8 | 27.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 1.8 | 47.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 1.7 | 23.9 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 1.7 | 11.8 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 1.6 | 4.9 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 1.6 | 35.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 1.6 | 28.6 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 1.6 | 23.6 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 1.6 | 20.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 1.5 | 4.6 | GO:0015734 | taurine transport(GO:0015734) |
| 1.5 | 3.0 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 1.5 | 8.9 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 1.5 | 32.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.4 | 17.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.4 | 4.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 1.4 | 31.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 1.4 | 9.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.3 | 13.2 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 1.3 | 10.5 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 1.3 | 5.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 1.3 | 6.4 | GO:0039516 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 1.3 | 5.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 1.2 | 12.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 1.2 | 9.8 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.1 | 29.6 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 1.1 | 18.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 1.1 | 12.8 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 1.1 | 1.1 | GO:0071650 | negative regulation of chemokine (C-C motif) ligand 5 production(GO:0071650) |
| 1.0 | 5.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.0 | 9.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 1.0 | 3.0 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 1.0 | 14.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 1.0 | 5.7 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 1.0 | 3.8 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.9 | 5.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.9 | 4.6 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.9 | 17.9 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.9 | 27.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.8 | 4.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.8 | 4.0 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.8 | 4.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.7 | 3.0 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.7 | 12.4 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.7 | 2.9 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.7 | 13.6 | GO:0060022 | hard palate development(GO:0060022) |
| 0.7 | 3.4 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.6 | 65.6 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.6 | 9.4 | GO:0015871 | choline transport(GO:0015871) negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.5 | 11.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.5 | 2.6 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.5 | 16.8 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.5 | 6.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.5 | 5.8 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.5 | 11.6 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.5 | 2.4 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.5 | 17.0 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.5 | 18.8 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.5 | 17.9 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.5 | 5.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.5 | 11.7 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.4 | 15.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.4 | 7.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.4 | 11.3 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.4 | 11.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 2.0 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.4 | 5.6 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.4 | 36.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.4 | 1.9 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.4 | 27.0 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.4 | 4.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.4 | 2.9 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.4 | 3.9 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.3 | 8.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.3 | 1.6 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.3 | 2.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 | 1.3 | GO:1902463 | actin filament branching(GO:0090135) protein localization to cell leading edge(GO:1902463) |
| 0.3 | 2.9 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.3 | 3.8 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.3 | 18.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.3 | 3.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 42.6 | GO:0002819 | regulation of adaptive immune response(GO:0002819) |
| 0.3 | 1.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.3 | 2.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 3.0 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.3 | 2.9 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.3 | 1.7 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.3 | 13.3 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.3 | 17.4 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.3 | 6.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 3.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.3 | 7.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 3.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 3.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 7.0 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.2 | 8.5 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.2 | 11.1 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.2 | 0.7 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.2 | 0.9 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.2 | 9.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.9 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.2 | 38.3 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 2.5 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 3.1 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.2 | 2.8 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.2 | 12.6 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.2 | 23.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 14.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 33.9 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.2 | 1.8 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.2 | 2.6 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 4.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.2 | 4.6 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.2 | 1.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 0.7 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.2 | 0.3 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 2.7 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 8.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 1.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 5.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 5.0 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 3.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 23.3 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 1.9 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 2.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 3.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 21.0 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 2.0 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.1 | 3.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 100.7 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.1 | 14.9 | GO:0034101 | erythrocyte homeostasis(GO:0034101) |
| 0.1 | 13.9 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 20.7 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.1 | 6.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 9.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.3 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 0.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 3.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.1 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.1 | 1.0 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 3.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.1 | 1.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 3.0 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.1 | 3.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 6.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.7 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.1 | 0.6 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 32.9 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 29.4 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 1.7 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 4.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.7 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 32.3 | GO:0006952 | defense response(GO:0006952) |
| 0.0 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 1.4 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.2 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 1.0 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.0 | 2.7 | GO:0072431 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 0.8 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 4.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 6.8 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 1.2 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 3.9 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 2.0 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 1.1 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 4.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 10.3 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 1.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.8 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 2.0 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.7 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 2.1 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.7 | 44.1 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 11.1 | 55.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 8.3 | 33.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 4.1 | 32.5 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 3.9 | 31.4 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 3.3 | 39.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 3.0 | 36.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 2.3 | 25.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 2.2 | 15.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 2.1 | 26.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.8 | 5.4 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 1.8 | 21.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 1.6 | 4.8 | GO:0031251 | PAN complex(GO:0031251) |
| 1.2 | 1.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 1.2 | 22.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 1.0 | 40.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 1.0 | 24.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.9 | 14.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.8 | 32.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.8 | 13.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.7 | 3.4 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.7 | 4.6 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.6 | 8.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.5 | 5.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.5 | 32.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.5 | 2.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.4 | 77.4 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.4 | 42.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.4 | 59.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.4 | 85.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.4 | 32.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.4 | 1.9 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.4 | 4.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.4 | 2.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.3 | 53.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.3 | 13.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.3 | 2.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 1.0 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.3 | 59.3 | GO:0016605 | PML body(GO:0016605) |
| 0.3 | 60.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.3 | 16.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 3.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 6.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.3 | 3.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.3 | 112.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.3 | 1.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 5.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 4.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 3.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 5.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 3.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 34.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 54.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 10.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 15.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.8 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 32.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 3.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 2.6 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 264.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 1.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 53.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 18.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 4.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 8.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 5.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 6.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 3.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 6.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 23.0 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.1 | 5.5 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 4.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.9 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.1 | 2.5 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.1 | 52.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 0.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 233.6 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 7.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 10.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 1.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.6 | 61.8 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 10.9 | 32.7 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 10.0 | 50.0 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 8.8 | 35.4 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 6.0 | 17.9 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 5.8 | 17.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 5.3 | 10.6 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 4.8 | 48.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 4.8 | 23.9 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 4.6 | 18.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 4.1 | 16.4 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 3.8 | 69.2 | GO:0019864 | IgG binding(GO:0019864) |
| 3.6 | 50.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 3.4 | 13.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 3.3 | 9.8 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 3.1 | 24.9 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 3.0 | 12.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 2.9 | 31.4 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 2.8 | 19.8 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 2.5 | 19.7 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 2.2 | 8.9 | GO:0001626 | nociceptin receptor activity(GO:0001626) |
| 2.0 | 16.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 2.0 | 47.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 1.8 | 25.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.8 | 56.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 1.7 | 28.6 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.6 | 16.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.6 | 9.4 | GO:0039552 | RIG-I binding(GO:0039552) |
| 1.5 | 4.6 | GO:0005369 | taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 1.5 | 23.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 1.5 | 124.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 1.5 | 10.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 1.4 | 4.1 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 1.2 | 34.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.2 | 64.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 1.2 | 9.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 1.1 | 11.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 1.1 | 15.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 1.1 | 39.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 1.0 | 25.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.0 | 3.0 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 1.0 | 10.8 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.9 | 5.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.9 | 4.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.8 | 16.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.8 | 11.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.8 | 5.4 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.8 | 13.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.7 | 6.8 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.7 | 19.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.6 | 35.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.6 | 2.5 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.6 | 9.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.6 | 56.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.5 | 2.7 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.5 | 4.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.5 | 12.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.5 | 11.7 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.5 | 11.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 5.6 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.5 | 66.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.4 | 2.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 3.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 5.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.4 | 23.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 23.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 3.8 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.3 | 14.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.3 | 15.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 19.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.3 | 139.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.3 | 6.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.3 | 3.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.3 | 4.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.3 | 3.4 | GO:0097100 | small ribosomal subunit rRNA binding(GO:0070181) supercoiled DNA binding(GO:0097100) |
| 0.2 | 2.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 3.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 5.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 8.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 3.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.2 | 7.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 23.1 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.2 | 9.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 10.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 4.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 4.6 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 100.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.2 | 3.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 2.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 2.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 10.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 3.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 5.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 13.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 15.8 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 65.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.9 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 2.9 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 2.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 9.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 3.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 15.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 1.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.3 | GO:0016436 | rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 4.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.8 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 1.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.0 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 4.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 7.2 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 9.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 8.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 37.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 3.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 14.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 3.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 1.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 3.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 2.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 17.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 1.2 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 3.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 2.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 7.7 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 7.0 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 1.7 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 48.0 | GO:0004872 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 3.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.6 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 80.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 1.2 | 137.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 1.2 | 74.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.1 | 103.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 1.0 | 36.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.9 | 57.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.8 | 72.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.8 | 55.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.5 | 17.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.5 | 28.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.4 | 11.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.4 | 48.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.4 | 18.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.4 | 49.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.4 | 11.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.4 | 4.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.3 | 16.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 9.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.3 | 18.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.3 | 24.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.3 | 28.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 6.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 9.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 15.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 37.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 55.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 4.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 5.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 89.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 2.5 | 39.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 2.0 | 46.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 1.8 | 48.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 1.5 | 103.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 1.3 | 50.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 1.3 | 25.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.2 | 24.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 1.1 | 24.9 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 1.0 | 60.5 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.8 | 108.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.8 | 29.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.7 | 34.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.6 | 11.8 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.6 | 5.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.6 | 58.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.5 | 6.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.5 | 56.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.4 | 32.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.4 | 33.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.4 | 127.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.4 | 16.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.4 | 33.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.4 | 6.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.4 | 13.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 5.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.3 | 13.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 14.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 4.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 2.7 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 6.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 21.6 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.2 | 5.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 24.9 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.2 | 4.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 32.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 4.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 3.6 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.2 | 4.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 31.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 3.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 13.9 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.1 | 3.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 14.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 3.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.1 | 3.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 28.8 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 2.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 2.2 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 1.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 4.7 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 3.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |