Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
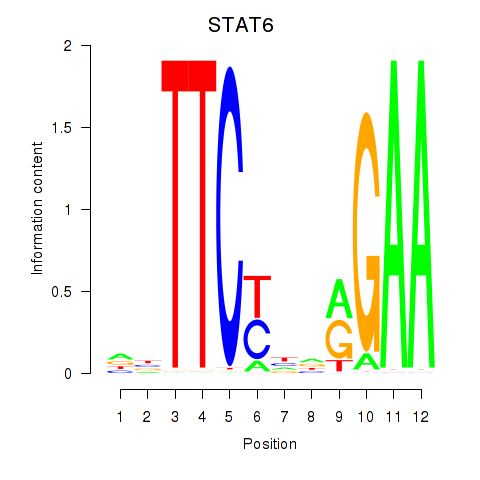
Results for STAT6
Z-value: 1.04
Transcription factors associated with STAT6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT6
|
ENSG00000166888.12 | signal transducer and activator of transcription 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT6 | hg38_v1_chr12_-_57111338_57111433 | -0.15 | 4.2e-01 | Click! |
Activity profile of STAT6 motif
Sorted Z-values of STAT6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_140784366 | 3.47 |
ENST00000674533.1
|
CDR1
|
cerebellar degeneration related protein 1 |
| chr18_-_26865689 | 3.32 |
ENST00000675739.1
ENST00000383168.9 ENST00000672981.2 ENST00000578776.1 |
AQP4
|
aquaporin 4 |
| chr14_-_24609660 | 2.90 |
ENST00000557220.6
ENST00000216338.9 ENST00000382548.4 |
GZMH
|
granzyme H |
| chr18_-_26865732 | 2.83 |
ENST00000672188.1
|
AQP4
|
aquaporin 4 |
| chr12_-_54973683 | 2.81 |
ENST00000532804.5
ENST00000531122.5 ENST00000533446.5 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr1_+_202462730 | 2.65 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr1_-_160647037 | 2.56 |
ENST00000302035.11
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr7_-_38300288 | 2.16 |
ENST00000390341.2
|
TRGV10
|
T cell receptor gamma variable 10 (non-functional) |
| chr1_-_160646958 | 2.10 |
ENST00000538290.2
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr2_+_190927649 | 2.04 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr17_+_36103819 | 1.97 |
ENST00000615863.2
ENST00000621626.1 |
CCL4
|
C-C motif chemokine ligand 4 |
| chr17_+_36211055 | 1.95 |
ENST00000617405.5
ENST00000617416.4 ENST00000613173.4 ENST00000620732.4 ENST00000620098.4 ENST00000620576.4 ENST00000620055.4 ENST00000610565.4 ENST00000620250.1 |
CCL4L2
|
C-C motif chemokine ligand 4 like 2 |
| chr12_-_54973547 | 1.88 |
ENST00000526532.5
ENST00000532757.5 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr5_-_84384871 | 1.73 |
ENST00000296591.10
|
EDIL3
|
EGF like repeats and discoidin domains 3 |
| chr7_+_80624961 | 1.71 |
ENST00000436384.5
|
CD36
|
CD36 molecule |
| chr14_-_24634160 | 1.70 |
ENST00000616551.1
ENST00000382542.5 ENST00000216341.9 ENST00000526004.1 ENST00000415355.7 |
GZMB
|
granzyme B |
| chr5_+_147878703 | 1.69 |
ENST00000296694.5
|
SCGB3A2
|
secretoglobin family 3A member 2 |
| chr5_+_119356011 | 1.69 |
ENST00000504771.3
ENST00000415806.2 |
TNFAIP8
|
TNF alpha induced protein 8 |
| chr12_-_110920568 | 1.66 |
ENST00000548438.1
ENST00000228841.15 |
MYL2
|
myosin light chain 2 |
| chr11_+_73647549 | 1.64 |
ENST00000227214.10
ENST00000398494.8 ENST00000543085.5 |
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr1_-_203229660 | 1.63 |
ENST00000255427.7
ENST00000367229.6 |
CHIT1
|
chitinase 1 |
| chr5_+_72179622 | 1.62 |
ENST00000504492.1
|
MAP1B
|
microtubule associated protein 1B |
| chr11_-_118212885 | 1.54 |
ENST00000524477.5
|
JAML
|
junction adhesion molecule like |
| chr8_-_81447428 | 1.53 |
ENST00000256103.3
ENST00000519260.1 |
PMP2
|
peripheral myelin protein 2 |
| chr2_-_60550900 | 1.52 |
ENST00000643222.1
ENST00000643459.1 ENST00000489516.7 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr14_+_22052503 | 1.46 |
ENST00000390449.3
|
TRAV21
|
T cell receptor alpha variable 21 |
| chr6_-_41286665 | 1.45 |
ENST00000589614.5
ENST00000244709.9 ENST00000334475.10 ENST00000591620.1 |
TREM1
|
triggering receptor expressed on myeloid cells 1 |
| chr15_+_91100194 | 1.39 |
ENST00000394232.6
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr5_+_36608146 | 1.38 |
ENST00000381918.4
ENST00000513646.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr3_+_159069252 | 1.35 |
ENST00000640015.1
ENST00000476809.7 ENST00000485419.7 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr3_+_50269140 | 1.34 |
ENST00000616701.5
ENST00000433753.4 ENST00000611067.4 |
SEMA3B
|
semaphorin 3B |
| chr16_+_30064731 | 1.29 |
ENST00000563987.5
|
ALDOA
|
aldolase, fructose-bisphosphate A |
| chr15_+_91099943 | 1.27 |
ENST00000545111.6
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chrX_+_136169624 | 1.25 |
ENST00000394153.6
|
FHL1
|
four and a half LIM domains 1 |
| chr16_-_29745951 | 1.25 |
ENST00000329410.4
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr3_+_121593363 | 1.24 |
ENST00000338040.6
|
FBXO40
|
F-box protein 40 |
| chr3_+_159273235 | 1.24 |
ENST00000638749.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr1_+_149782671 | 1.23 |
ENST00000444948.5
ENST00000369168.5 |
FCGR1A
|
Fc fragment of IgG receptor Ia |
| chr1_-_160647287 | 1.19 |
ENST00000235739.6
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr19_-_54100792 | 1.18 |
ENST00000391761.5
ENST00000356532.7 ENST00000616447.4 ENST00000359649.8 ENST00000358375.8 ENST00000391760.1 ENST00000351806.8 |
OSCAR
|
osteoclast associated Ig-like receptor |
| chr11_+_112961247 | 1.17 |
ENST00000621518.4
ENST00000618266.4 ENST00000615112.4 ENST00000615285.4 ENST00000619839.4 ENST00000401611.6 ENST00000621128.4 |
NCAM1
|
neural cell adhesion molecule 1 |
| chr7_-_38249572 | 1.16 |
ENST00000436911.6
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr9_-_133121228 | 1.14 |
ENST00000372050.8
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr17_-_35880350 | 1.11 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr17_+_36210924 | 1.08 |
ENST00000615418.4
|
CCL4L2
|
C-C motif chemokine ligand 4 like 2 |
| chrX_+_136169891 | 1.08 |
ENST00000449474.5
|
FHL1
|
four and a half LIM domains 1 |
| chr15_+_66453418 | 1.07 |
ENST00000566326.1
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chrX_+_136169664 | 1.07 |
ENST00000456445.5
|
FHL1
|
four and a half LIM domains 1 |
| chrX_+_136169833 | 1.05 |
ENST00000628032.2
|
FHL1
|
four and a half LIM domains 1 |
| chr14_-_24634266 | 1.05 |
ENST00000382540.5
|
GZMB
|
granzyme B |
| chr5_-_88823763 | 1.03 |
ENST00000635898.1
ENST00000626391.2 ENST00000628656.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr4_+_143391506 | 0.98 |
ENST00000509992.1
|
GAB1
|
GRB2 associated binding protein 1 |
| chr11_+_112961480 | 0.97 |
ENST00000621850.4
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr22_+_32043253 | 0.96 |
ENST00000266088.9
|
SLC5A1
|
solute carrier family 5 member 1 |
| chr17_+_38925168 | 0.96 |
ENST00000583195.2
|
LINC00672
|
long intergenic non-protein coding RNA 672 |
| chr17_-_8867639 | 0.95 |
ENST00000619866.5
|
PIK3R6
|
phosphoinositide-3-kinase regulatory subunit 6 |
| chr20_+_59676661 | 0.95 |
ENST00000355648.8
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr19_-_7702139 | 0.95 |
ENST00000346664.9
|
FCER2
|
Fc fragment of IgE receptor II |
| chr1_+_192636121 | 0.94 |
ENST00000543215.5
ENST00000391995.7 |
RGS13
|
regulator of G protein signaling 13 |
| chr19_-_7702124 | 0.93 |
ENST00000597921.6
|
FCER2
|
Fc fragment of IgE receptor II |
| chr11_+_112961402 | 0.93 |
ENST00000613217.4
ENST00000316851.12 ENST00000620046.4 ENST00000531044.5 ENST00000529356.5 |
NCAM1
|
neural cell adhesion molecule 1 |
| chr11_-_128587551 | 0.93 |
ENST00000392668.8
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr16_+_30064462 | 0.93 |
ENST00000412304.6
|
ALDOA
|
aldolase, fructose-bisphosphate A |
| chr7_-_38265678 | 0.93 |
ENST00000443402.6
|
TRGC1
|
T cell receptor gamma constant 1 |
| chr2_+_68734861 | 0.92 |
ENST00000467265.5
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr13_+_48653921 | 0.92 |
ENST00000682523.1
|
CYSLTR2
|
cysteinyl leukotriene receptor 2 |
| chr19_-_7700074 | 0.91 |
ENST00000593418.1
|
FCER2
|
Fc fragment of IgE receptor II |
| chr2_-_174597795 | 0.90 |
ENST00000679041.1
|
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr2_+_68734773 | 0.90 |
ENST00000409202.8
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr12_+_57578238 | 0.90 |
ENST00000675907.1
|
KIF5A
|
kinesin family member 5A |
| chr6_-_24877262 | 0.89 |
ENST00000378023.8
ENST00000540914.5 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr22_+_44069043 | 0.89 |
ENST00000404989.1
|
PARVB
|
parvin beta |
| chr17_+_34255274 | 0.88 |
ENST00000580907.5
ENST00000225831.4 |
CCL2
|
C-C motif chemokine ligand 2 |
| chr2_-_174597728 | 0.88 |
ENST00000409891.5
ENST00000410117.5 |
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr4_-_46993520 | 0.87 |
ENST00000264318.4
|
GABRA4
|
gamma-aminobutyric acid type A receptor subunit alpha4 |
| chr11_-_59212869 | 0.85 |
ENST00000361050.4
|
MPEG1
|
macrophage expressed 1 |
| chr7_-_139716980 | 0.84 |
ENST00000342645.7
|
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr6_+_31572279 | 0.82 |
ENST00000418386.3
|
LTA
|
lymphotoxin alpha |
| chr1_-_35554930 | 0.80 |
ENST00000440579.5
ENST00000494948.1 |
KIAA0319L
|
KIAA0319 like |
| chr1_+_121087343 | 0.80 |
ENST00000616817.4
ENST00000623603.3 ENST00000369384.9 ENST00000369383.8 ENST00000369178.5 |
FCGR1B
|
Fc fragment of IgG receptor Ib |
| chr6_-_32853618 | 0.79 |
ENST00000354258.5
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr6_-_41201128 | 0.79 |
ENST00000483722.2
|
TREML2
|
triggering receptor expressed on myeloid cells like 2 |
| chr2_-_201451446 | 0.78 |
ENST00000332624.8
ENST00000430254.1 |
TRAK2
|
trafficking kinesin protein 2 |
| chr18_+_13277351 | 0.78 |
ENST00000679091.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr18_+_68715191 | 0.78 |
ENST00000578970.5
ENST00000582371.5 ENST00000584775.5 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr10_-_132307935 | 0.78 |
ENST00000298630.8
|
STK32C
|
serine/threonine kinase 32C |
| chr12_-_54385727 | 0.78 |
ENST00000551109.5
ENST00000546970.5 |
ZNF385A
|
zinc finger protein 385A |
| chr15_+_24954912 | 0.76 |
ENST00000584968.5
ENST00000346403.10 ENST00000554227.6 ENST00000390687.9 ENST00000579070.5 ENST00000577565.1 ENST00000577949.5 ENST00000338327.4 |
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N SNRPN upstream reading frame |
| chr6_-_32853813 | 0.73 |
ENST00000643049.2
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr7_-_38929550 | 0.73 |
ENST00000418457.6
|
VPS41
|
VPS41 subunit of HOPS complex |
| chr2_-_133568393 | 0.73 |
ENST00000317721.10
ENST00000405974.7 ENST00000409261.6 ENST00000409213.5 |
NCKAP5
|
NCK associated protein 5 |
| chr16_+_30064142 | 0.72 |
ENST00000562168.5
ENST00000569545.5 |
ALDOA
|
aldolase, fructose-bisphosphate A |
| chr11_+_65314853 | 0.68 |
ENST00000279249.3
|
CDC42EP2
|
CDC42 effector protein 2 |
| chrX_-_73214793 | 0.68 |
ENST00000373517.4
|
NAP1L2
|
nucleosome assembly protein 1 like 2 |
| chr16_+_30064274 | 0.68 |
ENST00000563060.6
|
ALDOA
|
aldolase, fructose-bisphosphate A |
| chr5_-_116554858 | 0.67 |
ENST00000509665.1
|
SEMA6A
|
semaphorin 6A |
| chr12_-_118052597 | 0.67 |
ENST00000535496.5
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chrX_-_53281524 | 0.67 |
ENST00000498281.2
|
IQSEC2
|
IQ motif and Sec7 domain ArfGEF 2 |
| chr5_-_147081428 | 0.66 |
ENST00000394413.7
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr3_-_98517096 | 0.66 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr14_+_21918161 | 0.65 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chrX_-_50643649 | 0.65 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chr4_+_30720348 | 0.64 |
ENST00000361762.3
|
PCDH7
|
protocadherin 7 |
| chr20_-_23086316 | 0.62 |
ENST00000246006.5
|
CD93
|
CD93 molecule |
| chr2_+_106487349 | 0.61 |
ENST00000643224.2
ENST00000416057.2 |
CD8B2
|
CD8b2 molecule |
| chr11_-_790062 | 0.61 |
ENST00000330106.5
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr5_+_127649018 | 0.60 |
ENST00000379445.7
|
CTXN3
|
cortexin 3 |
| chr5_+_35856883 | 0.60 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr3_-_179974254 | 0.59 |
ENST00000468741.5
|
PEX5L
|
peroxisomal biogenesis factor 5 like |
| chr8_-_142777174 | 0.57 |
ENST00000652477.1
ENST00000614491.1 ENST00000613110.4 |
LYNX1
|
Ly6/neurotoxin 1 |
| chr4_+_6693870 | 0.57 |
ENST00000296370.4
|
S100P
|
S100 calcium binding protein P |
| chr2_-_151971750 | 0.55 |
ENST00000636598.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr6_+_26440472 | 0.54 |
ENST00000494393.5
ENST00000482451.5 ENST00000471353.5 ENST00000361232.7 ENST00000487627.5 ENST00000496719.1 ENST00000244519.7 ENST00000490254.5 ENST00000487272.1 |
BTN3A3
|
butyrophilin subfamily 3 member A3 |
| chr6_+_31571957 | 0.54 |
ENST00000454783.5
|
LTA
|
lymphotoxin alpha |
| chr3_+_141402322 | 0.53 |
ENST00000510338.5
ENST00000504673.5 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr12_-_46269951 | 0.52 |
ENST00000550173.1
|
SLC38A1
|
solute carrier family 38 member 1 |
| chr16_-_68000549 | 0.52 |
ENST00000575510.5
|
DPEP2
|
dipeptidase 2 |
| chr12_+_94262521 | 0.51 |
ENST00000545312.1
|
PLXNC1
|
plexin C1 |
| chr12_+_8092881 | 0.51 |
ENST00000638334.1
|
NECAP1
|
NECAP endocytosis associated 1 |
| chr4_-_120066777 | 0.50 |
ENST00000296509.11
|
MAD2L1
|
mitotic arrest deficient 2 like 1 |
| chr8_-_116874528 | 0.50 |
ENST00000517485.5
|
RAD21
|
RAD21 cohesin complex component |
| chr5_-_74866958 | 0.49 |
ENST00000389156.9
|
FAM169A
|
family with sequence similarity 169 member A |
| chr15_+_77015811 | 0.48 |
ENST00000559859.5
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr12_+_8992029 | 0.47 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr6_+_32854179 | 0.45 |
ENST00000374859.3
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr2_+_167248638 | 0.45 |
ENST00000295237.10
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr11_+_118359572 | 0.45 |
ENST00000252108.8
ENST00000431736.6 |
UBE4A
|
ubiquitination factor E4A |
| chr5_+_139273752 | 0.44 |
ENST00000509990.5
ENST00000506147.5 ENST00000512107.5 |
MATR3
|
matrin 3 |
| chr19_-_42442938 | 0.44 |
ENST00000601181.6
|
CXCL17
|
C-X-C motif chemokine ligand 17 |
| chr6_-_33787001 | 0.43 |
ENST00000508327.5
ENST00000513701.1 |
LEMD2
|
LEM domain nuclear envelope protein 2 |
| chr18_-_55401525 | 0.42 |
ENST00000562030.3
ENST00000569012.5 |
TCF4
|
transcription factor 4 |
| chr5_-_141958174 | 0.42 |
ENST00000231484.4
|
PCDH12
|
protocadherin 12 |
| chr5_+_94618653 | 0.42 |
ENST00000265140.10
ENST00000504099.5 |
SLF1
|
SMC5-SMC6 complex localization factor 1 |
| chr2_-_119366757 | 0.42 |
ENST00000414534.1
|
C2orf76
|
chromosome 2 open reading frame 76 |
| chr13_+_48653711 | 0.41 |
ENST00000614739.4
ENST00000617562.4 ENST00000621321.1 ENST00000622559.4 |
CYSLTR2
|
cysteinyl leukotriene receptor 2 |
| chr9_-_10612703 | 0.41 |
ENST00000463477.5
|
PTPRD
|
protein tyrosine phosphatase receptor type D |
| chr4_-_48114523 | 0.41 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr1_-_100178215 | 0.41 |
ENST00000370138.1
ENST00000370137.6 ENST00000342895.7 ENST00000620882.4 |
LRRC39
|
leucine rich repeat containing 39 |
| chr5_-_74866914 | 0.40 |
ENST00000513277.5
|
FAM169A
|
family with sequence similarity 169 member A |
| chr12_-_110920710 | 0.40 |
ENST00000546404.1
|
MYL2
|
myosin light chain 2 |
| chr8_-_116874746 | 0.40 |
ENST00000297338.7
|
RAD21
|
RAD21 cohesin complex component |
| chr4_+_112860981 | 0.40 |
ENST00000671704.1
|
ANK2
|
ankyrin 2 |
| chr2_-_108989206 | 0.39 |
ENST00000258443.7
ENST00000409271.5 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr4_+_112860912 | 0.39 |
ENST00000671951.1
|
ANK2
|
ankyrin 2 |
| chr4_+_159241016 | 0.38 |
ENST00000644902.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr14_+_55661242 | 0.38 |
ENST00000553624.5
|
KTN1
|
kinectin 1 |
| chr17_-_39778213 | 0.38 |
ENST00000583368.1
|
IKZF3
|
IKAROS family zinc finger 3 |
| chr4_-_46390100 | 0.37 |
ENST00000381620.9
|
GABRA2
|
gamma-aminobutyric acid type A receptor subunit alpha2 |
| chr5_-_88824266 | 0.37 |
ENST00000509373.1
ENST00000636541.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr22_-_41947087 | 0.37 |
ENST00000407253.7
ENST00000215980.10 |
CENPM
|
centromere protein M |
| chr11_+_8019193 | 0.36 |
ENST00000534099.5
|
TUB
|
TUB bipartite transcription factor |
| chr5_-_147081462 | 0.36 |
ENST00000508267.5
ENST00000504198.5 |
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr1_-_45499931 | 0.36 |
ENST00000626657.2
|
CCDC163
|
coiled-coil domain containing 163 |
| chr9_-_132079856 | 0.36 |
ENST00000651555.1
ENST00000651950.1 ENST00000357028.6 ENST00000474263.1 ENST00000292035.10 |
MED27
|
mediator complex subunit 27 |
| chr2_+_12716893 | 0.35 |
ENST00000381465.2
ENST00000155926.9 |
TRIB2
|
tribbles pseudokinase 2 |
| chr4_+_112861053 | 0.35 |
ENST00000672221.1
|
ANK2
|
ankyrin 2 |
| chr20_+_43945677 | 0.34 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr5_-_151686953 | 0.34 |
ENST00000538026.5
ENST00000522348.1 ENST00000521569.1 |
SPARC
|
secreted protein acidic and cysteine rich |
| chr10_-_59362460 | 0.31 |
ENST00000422313.6
ENST00000435852.6 ENST00000614220.4 ENST00000618804.5 ENST00000621119.4 |
FAM13C
|
family with sequence similarity 13 member C |
| chr4_-_46390273 | 0.31 |
ENST00000515082.5
|
GABRA2
|
gamma-aminobutyric acid type A receptor subunit alpha2 |
| chr13_+_50015438 | 0.30 |
ENST00000312942.2
|
KCNRG
|
potassium channel regulator |
| chr4_-_46390039 | 0.30 |
ENST00000540012.5
|
GABRA2
|
gamma-aminobutyric acid type A receptor subunit alpha2 |
| chr1_-_45500040 | 0.30 |
ENST00000629482.3
|
CCDC163
|
coiled-coil domain containing 163 |
| chr11_-_60952559 | 0.30 |
ENST00000538739.2
|
SLC15A3
|
solute carrier family 15 member 3 |
| chr11_-_72752376 | 0.29 |
ENST00000393609.8
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr1_+_150272772 | 0.29 |
ENST00000369098.3
ENST00000369099.8 |
C1orf54
|
chromosome 1 open reading frame 54 |
| chr11_-_77820706 | 0.28 |
ENST00000440064.2
ENST00000528095.5 ENST00000308488.11 |
RSF1
|
remodeling and spacing factor 1 |
| chr2_+_113127588 | 0.28 |
ENST00000409930.4
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr3_+_141262614 | 0.28 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chr15_+_78873723 | 0.28 |
ENST00000559690.5
ENST00000559158.5 |
MORF4L1
|
mortality factor 4 like 1 |
| chr20_+_59958422 | 0.27 |
ENST00000348616.9
|
CDH26
|
cadherin 26 |
| chr6_+_30557274 | 0.27 |
ENST00000376557.3
|
PRR3
|
proline rich 3 |
| chr12_-_124495252 | 0.27 |
ENST00000405201.5
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr11_+_77821125 | 0.26 |
ENST00000526415.5
ENST00000393427.6 ENST00000527134.5 ENST00000304716.12 ENST00000630098.2 |
AAMDC
|
adipogenesis associated Mth938 domain containing |
| chr5_-_151686908 | 0.26 |
ENST00000231061.9
|
SPARC
|
secreted protein acidic and cysteine rich |
| chr22_-_41946688 | 0.26 |
ENST00000404067.5
ENST00000402338.5 |
CENPM
|
centromere protein M |
| chr13_-_51845169 | 0.26 |
ENST00000627246.3
ENST00000629372.3 |
TMEM272
|
transmembrane protein 272 |
| chr11_+_65333834 | 0.25 |
ENST00000528416.6
ENST00000415073.6 ENST00000252268.8 |
DPF2
|
double PHD fingers 2 |
| chr1_-_108192818 | 0.25 |
ENST00000370041.4
|
SLC25A24
|
solute carrier family 25 member 24 |
| chr13_+_50015254 | 0.24 |
ENST00000360473.8
|
KCNRG
|
potassium channel regulator |
| chr2_-_229921903 | 0.23 |
ENST00000389045.7
ENST00000409677.5 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr14_+_38207893 | 0.22 |
ENST00000267377.3
|
SSTR1
|
somatostatin receptor 1 |
| chr2_-_229921316 | 0.22 |
ENST00000428959.5
ENST00000675423.1 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr3_-_49813880 | 0.22 |
ENST00000333486.4
|
UBA7
|
ubiquitin like modifier activating enzyme 7 |
| chr13_-_29586788 | 0.22 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 member 1 |
| chr11_+_63888515 | 0.21 |
ENST00000509502.6
ENST00000512060.1 |
MARK2
|
microtubule affinity regulating kinase 2 |
| chr1_-_190477854 | 0.21 |
ENST00000367462.5
|
BRINP3
|
BMP/retinoic acid inducible neural specific 3 |
| chr12_-_81759307 | 0.21 |
ENST00000547623.5
ENST00000549396.6 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr1_+_77918128 | 0.20 |
ENST00000342754.5
|
NEXN
|
nexilin F-actin binding protein |
| chr1_+_66330711 | 0.20 |
ENST00000528771.5
|
PDE4B
|
phosphodiesterase 4B |
| chr14_+_55661272 | 0.20 |
ENST00000555573.5
|
KTN1
|
kinectin 1 |
| chr2_+_131011683 | 0.19 |
ENST00000355771.7
|
ARHGEF4
|
Rho guanine nucleotide exchange factor 4 |
| chr10_-_59362584 | 0.19 |
ENST00000618427.4
ENST00000611933.4 |
FAM13C
|
family with sequence similarity 13 member C |
| chr8_+_122781621 | 0.19 |
ENST00000314393.6
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr3_+_130443484 | 0.18 |
ENST00000512482.1
|
COL6A5
|
collagen type VI alpha 5 chain |
| chr12_-_81758641 | 0.18 |
ENST00000552948.5
ENST00000548586.5 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr12_+_56041893 | 0.18 |
ENST00000552361.1
ENST00000646449.2 |
RPS26
|
ribosomal protein S26 |
| chr9_-_7799752 | 0.17 |
ENST00000358227.5
|
DMAC1
|
distal membrane arm assembly complex 1 |
| chr7_-_77199808 | 0.17 |
ENST00000248598.6
|
FGL2
|
fibrinogen like 2 |
| chr1_+_213989691 | 0.17 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.8 | GO:0035744 | T-helper 1 cell cytokine production(GO:0035744) |
| 0.8 | 4.7 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.6 | 3.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.5 | 4.2 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.5 | 1.5 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.3 | 2.7 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.3 | 0.9 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.3 | 2.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 1.7 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.3 | 1.5 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.2 | 0.7 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.2 | 1.6 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 1.4 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.2 | 1.5 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.2 | 3.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.5 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.2 | 2.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 6.2 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.9 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.1 | 1.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 1.8 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 1.7 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 1.6 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 1.0 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.1 | 1.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 0.8 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.4 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 1.1 | GO:0098907 | protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.1 | 0.6 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.7 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.9 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.9 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 0.9 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 0.6 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 0.3 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.4 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.5 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 1.6 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.1 | 0.6 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.2 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.4 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.6 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 4.5 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.5 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.2 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 2.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.5 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.4 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 5.1 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.4 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:1904863 | regulation of beta-catenin-TCF complex assembly(GO:1904863) negative regulation of beta-catenin-TCF complex assembly(GO:1904864) |
| 0.0 | 0.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.4 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 1.0 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:1904744 | positive regulation of telomeric DNA binding(GO:1904744) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 1.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 1.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0042269 | regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.0 | 0.9 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 1.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.5 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.6 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 1.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 2.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.7 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.9 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 0.9 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 6.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 2.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 4.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.8 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 2.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 18.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.8 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 1.9 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 2.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.3 | 6.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 2.8 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 3.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 2.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 0.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 0.6 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 1.5 | GO:0046979 | MHC class Ib protein binding(GO:0023029) TAP2 binding(GO:0046979) |
| 0.2 | 1.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 1.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.8 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 1.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 1.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 2.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.6 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 7.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 1.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 2.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.2 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 1.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 3.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.0 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 3.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 4.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 4.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 2.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 4.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 2.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 4.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 3.0 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 2.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 2.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.6 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 2.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |