Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
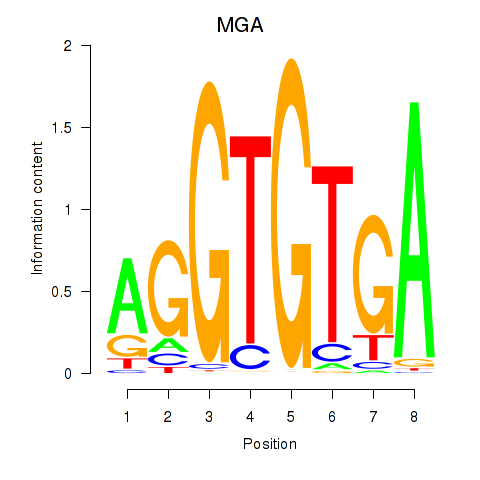
Results for TBX15_MGA
Z-value: 1.37
Transcription factors associated with TBX15_MGA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
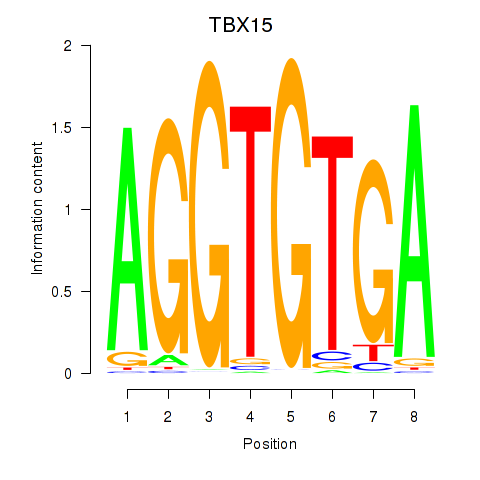
TBX15
|
ENSG00000092607.15 | T-box transcription factor 15 |
|
MGA
|
ENSG00000174197.17 | MAX dimerization protein MGA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MGA | hg38_v1_chr15_+_41660397_41660471 | -0.15 | 4.2e-01 | Click! |
| TBX15 | hg38_v1_chr1_-_118989504_118989556 | 0.03 | 8.6e-01 | Click! |
Activity profile of TBX15_MGA motif
Sorted Z-values of TBX15_MGA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX15_MGA
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 1.2 | 4.7 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 1.0 | 3.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 1.0 | 4.8 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.9 | 2.8 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.9 | 3.5 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.8 | 4.2 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.8 | 2.3 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.7 | 2.2 | GO:1990641 | response to iron ion starvation(GO:1990641) |
| 0.7 | 3.7 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.7 | 2.2 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.7 | 2.8 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.6 | 2.5 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.6 | 1.9 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.6 | 1.8 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.6 | 2.8 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.6 | 1.7 | GO:0090381 | cardiac cell fate determination(GO:0060913) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.5 | 2.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.5 | 2.9 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.5 | 1.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.5 | 1.4 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.4 | 1.2 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.4 | 2.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.4 | 1.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 2.5 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.3 | 2.0 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.3 | 2.3 | GO:0030421 | defecation(GO:0030421) |
| 0.3 | 4.3 | GO:0015820 | leucine transport(GO:0015820) |
| 0.3 | 5.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.3 | 2.6 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.3 | 1.3 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.3 | 1.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.3 | 6.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.3 | 0.9 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.3 | 0.9 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.3 | 1.2 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.3 | 1.8 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 2.8 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.3 | 1.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.3 | 2.6 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.3 | 1.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 | 7.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 5.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 6.5 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.2 | 1.4 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.2 | 0.7 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 1.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.3 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.2 | 0.9 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.2 | 1.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.2 | 3.5 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.2 | 0.6 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.2 | 1.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 2.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 1.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 1.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 1.0 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.2 | 1.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.2 | 1.2 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.2 | 0.6 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) regulation of mRNA modification(GO:0090365) |
| 0.2 | 0.9 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.2 | 0.6 | GO:0015882 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 | 1.4 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 0.5 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 | 0.7 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.2 | 1.5 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 1.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.2 | 4.5 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 2.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.2 | 3.4 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.2 | 0.9 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.2 | 1.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 0.6 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.1 | 0.7 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 4.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.6 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.6 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 1.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 14.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.5 | GO:1904327 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 1.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 2.4 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.8 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.9 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 7.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.4 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.6 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 1.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.5 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 1.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.4 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 1.8 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 1.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.3 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 2.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:2000592 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney field specification(GO:0072004) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 3.0 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 3.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 1.9 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.5 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 1.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 5.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.4 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 0.5 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 1.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 2.5 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 0.4 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 1.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.2 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 1.7 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.1 | 0.2 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.1 | 0.6 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 1.4 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.9 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.7 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.1 | 1.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 1.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 4.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.5 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 1.5 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.1 | 1.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.9 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 2.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.9 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.6 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.1 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 1.3 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 | 0.4 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.4 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.7 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) |
| 0.1 | 0.2 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.1 | 2.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.4 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 1.0 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 2.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 2.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.3 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 3.1 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 2.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 0.4 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.1 | 1.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 1.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 1.2 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 1.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.5 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.7 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.2 | GO:0036265 | peptidyl-glutamine methylation(GO:0018364) RNA (guanine-N7)-methylation(GO:0036265) rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 | 0.4 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 1.4 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 2.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 1.3 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 2.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.3 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 3.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 1.0 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 1.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.3 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.4 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 2.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.2 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0030719 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.0 | 0.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 4.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.2 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.4 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 5.2 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.7 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 2.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.2 | GO:1903588 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 1.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 2.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.3 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 1.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.3 | GO:0046885 | regulation of hormone biosynthetic process(GO:0046885) |
| 0.0 | 0.8 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.2 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 1.0 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 2.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 2.0 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.6 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.2 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.2 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 1.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 1.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.1 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 1.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.8 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.6 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.8 | GO:2000757 | negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.6 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.0 | 1.1 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.2 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.5 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 0.0 | 0.2 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 1.3 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.9 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 1.3 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 1.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 1.2 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) |
| 0.0 | 0.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 4.9 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 2.0 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.1 | GO:0006023 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.3 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 5.8 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.3 | 2.0 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 1.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.3 | 5.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 2.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 1.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 6.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 1.7 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 10.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 0.9 | GO:1990742 | microvesicle(GO:1990742) |
| 0.2 | 1.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 3.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 1.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 2.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.2 | 7.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 4.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.8 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 1.5 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 2.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 5.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 5.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 15.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.3 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 1.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 2.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.5 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.1 | 0.3 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) translation initiation ternary complex(GO:0044207) |
| 0.1 | 2.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 5.6 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 3.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.9 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 6.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.7 | GO:0044326 | chromaffin granule membrane(GO:0042584) dendritic spine neck(GO:0044326) |
| 0.1 | 1.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 2.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 4.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 16.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.9 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 5.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 4.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 3.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.6 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 2.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 1.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 15.3 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 2.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.2 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 3.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0097545 | axonemal outer doublet(GO:0097545) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 2.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.7 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 2.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.0 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 1.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 5.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 6.3 | GO:0098794 | postsynapse(GO:0098794) |
| 0.0 | 0.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.0 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.9 | 2.8 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.6 | 1.9 | GO:0061599 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.6 | 5.4 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.6 | 1.8 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.6 | 2.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.5 | 3.3 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.5 | 7.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.5 | 1.8 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.5 | 1.8 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.4 | 2.6 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.4 | 1.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.4 | 1.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.4 | 3.8 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.4 | 1.2 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.4 | 2.0 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.4 | 1.2 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.4 | 2.3 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.3 | 8.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.3 | 3.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.3 | 1.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.3 | 1.4 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.3 | 0.8 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.3 | 4.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 4.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 2.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 3.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 2.4 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 3.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 0.7 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 0.9 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 3.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 0.6 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.2 | 2.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 1.9 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 5.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 6.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 1.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 3.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 0.6 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.2 | 0.9 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 0.9 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 8.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 2.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 4.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 9.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 1.0 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 1.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 3.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 3.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 1.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 7.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 1.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.5 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 1.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.5 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.1 | 1.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 1.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.5 | GO:0005115 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 0.3 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.9 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 17.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.4 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 2.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 6.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 2.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 2.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.3 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.1 | 4.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.2 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 2.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.9 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 2.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.8 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 2.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.2 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.1 | 1.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 1.0 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 2.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.9 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 1.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.5 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 1.6 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 1.0 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 1.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.6 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.2 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 2.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 3.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 3-dioxygenase activity(GO:0019797) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 2.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 3.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 2.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.0 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 2.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0005416 | cation:amino acid symporter activity(GO:0005416) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0008066 | glutamate receptor activity(GO:0008066) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 10.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 4.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 2.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 4.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 1.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 4.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 3.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 4.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 15.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 4.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 6.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 7.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 6.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 4.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 5.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 3.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 5.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 3.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 2.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 4.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 5.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 1.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.6 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 1.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 6.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |