Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for TBX20
Z-value: 0.82
Transcription factors associated with TBX20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX20
|
ENSG00000164532.11 | T-box transcription factor 20 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX20 | hg38_v1_chr7_-_35254074_35254106 | -0.23 | 2.1e-01 | Click! |
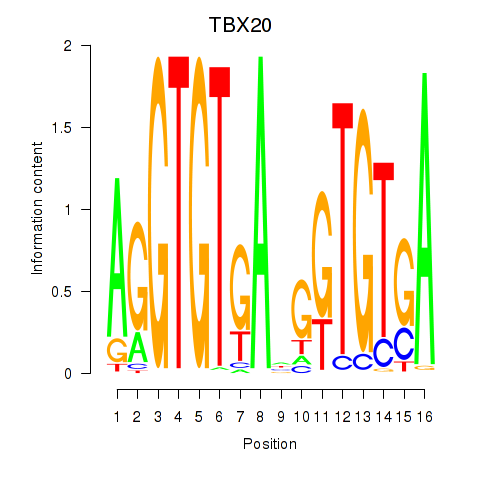
Activity profile of TBX20 motif
Sorted Z-values of TBX20 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_57628507 | 17.01 |
ENST00000456916.5
ENST00000567154.5 ENST00000388813.9 ENST00000562558.6 ENST00000566271.6 ENST00000563374.5 ENST00000673126.2 ENST00000562631.7 ENST00000568234.5 ENST00000565770.5 ENST00000566164.5 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr16_+_57628684 | 11.98 |
ENST00000567397.5
ENST00000568979.5 ENST00000672974.1 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr8_+_27325516 | 2.78 |
ENST00000346049.10
ENST00000420218.3 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr15_-_22160868 | 2.68 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chrX_+_78945332 | 2.32 |
ENST00000544091.1
ENST00000171757.3 |
P2RY10
|
P2Y receptor family member 10 |
| chr15_-_21718245 | 2.12 |
ENST00000630556.1
|
ENSG00000281179.1
|
novel gene identicle to IGHV1OR15-1 |
| chr14_-_105987068 | 2.00 |
ENST00000390594.3
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr12_+_15322480 | 1.66 |
ENST00000674188.1
ENST00000281171.9 ENST00000543886.6 |
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr14_-_106235582 | 1.49 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr6_-_2841853 | 1.35 |
ENST00000380739.6
|
SERPINB1
|
serpin family B member 1 |
| chr5_+_134525649 | 1.28 |
ENST00000282605.8
ENST00000681547.2 ENST00000361895.6 ENST00000402835.5 |
JADE2
|
jade family PHD finger 2 |
| chrX_+_68693629 | 1.15 |
ENST00000374597.3
|
STARD8
|
StAR related lipid transfer domain containing 8 |
| chr7_+_30145789 | 1.14 |
ENST00000324489.5
|
MTURN
|
maturin, neural progenitor differentiation regulator homolog |
| chr12_+_15322257 | 1.07 |
ENST00000674316.1
|
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr12_-_121038967 | 1.00 |
ENST00000680620.1
ENST00000679655.1 ENST00000543677.2 |
OASL
|
2'-5'-oligoadenylate synthetase like |
| chr17_-_3696133 | 0.99 |
ENST00000225328.10
|
P2RX5
|
purinergic receptor P2X 5 |
| chr7_+_142424942 | 0.98 |
ENST00000426318.2
|
TRBV10-2
|
T cell receptor beta variable 10-2 |
| chr19_-_39320839 | 0.95 |
ENST00000248668.5
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr3_-_183555696 | 0.93 |
ENST00000341319.8
|
KLHL6
|
kelch like family member 6 |
| chr12_+_15322529 | 0.93 |
ENST00000348962.7
|
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr16_+_31033781 | 0.92 |
ENST00000613541.1
|
STX4
|
syntaxin 4 |
| chr4_+_159104148 | 0.84 |
ENST00000505478.5
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr10_-_54801262 | 0.78 |
ENST00000373965.6
ENST00000616114.4 ENST00000621708.4 ENST00000495484.5 ENST00000395440.5 ENST00000395442.5 ENST00000617271.4 ENST00000395446.5 ENST00000409834.5 ENST00000361849.7 ENST00000373957.7 ENST00000395430.5 ENST00000395433.5 ENST00000437009.5 |
PCDH15
|
protocadherin related 15 |
| chr2_+_60881553 | 0.77 |
ENST00000394479.4
|
REL
|
REL proto-oncogene, NF-kB subunit |
| chr12_-_121039236 | 0.71 |
ENST00000257570.9
|
OASL
|
2'-5'-oligoadenylate synthetase like |
| chr12_-_121039204 | 0.70 |
ENST00000620239.5
|
OASL
|
2'-5'-oligoadenylate synthetase like |
| chr16_+_31033513 | 0.67 |
ENST00000313843.8
|
STX4
|
syntaxin 4 |
| chr5_+_162067500 | 0.67 |
ENST00000639384.1
ENST00000640985.1 ENST00000638772.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr2_+_60881515 | 0.66 |
ENST00000295025.12
|
REL
|
REL proto-oncogene, NF-kB subunit |
| chr12_-_121039156 | 0.65 |
ENST00000339275.10
|
OASL
|
2'-5'-oligoadenylate synthetase like |
| chr5_+_136059151 | 0.62 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor beta induced |
| chr5_+_162067458 | 0.62 |
ENST00000639975.1
ENST00000639111.2 ENST00000639683.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr10_-_102114935 | 0.62 |
ENST00000361198.9
|
LDB1
|
LIM domain binding 1 |
| chr18_-_55587335 | 0.62 |
ENST00000638154.3
|
TCF4
|
transcription factor 4 |
| chrX_-_101617921 | 0.51 |
ENST00000361910.9
ENST00000538627.5 ENST00000539247.5 |
ARMCX6
|
armadillo repeat containing X-linked 6 |
| chr12_-_16609869 | 0.45 |
ENST00000534946.5
|
LMO3
|
LIM domain only 3 |
| chr1_+_87331668 | 0.44 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr19_+_7680822 | 0.43 |
ENST00000596148.3
ENST00000317378.5 ENST00000426877.2 |
TRAPPC5
|
trafficking protein particle complex 5 |
| chr12_-_16610037 | 0.41 |
ENST00000540590.1
|
LMO3
|
LIM domain only 3 |
| chr3_+_187197486 | 0.36 |
ENST00000312295.5
|
RTP1
|
receptor transporter protein 1 |
| chr13_+_113001270 | 0.30 |
ENST00000409954.6
|
MCF2L
|
MCF.2 cell line derived transforming sequence like |
| chr3_-_50359480 | 0.28 |
ENST00000266025.4
|
TMEM115
|
transmembrane protein 115 |
| chr19_+_45668191 | 0.25 |
ENST00000652180.1
ENST00000590918.6 ENST00000263281.7 ENST00000304207.12 |
GIPR
|
gastric inhibitory polypeptide receptor |
| chr8_-_141496759 | 0.23 |
ENST00000521161.5
|
MROH5
|
maestro heat like repeat family member 5 (gene/pseudogene) |
| chr8_-_108443409 | 0.20 |
ENST00000678023.1
ENST00000679198.1 ENST00000678881.1 ENST00000677084.1 ENST00000676663.1 ENST00000677614.1 ENST00000677040.1 ENST00000676487.1 ENST00000677409.1 ENST00000678797.1 ENST00000678334.1 |
EIF3E
|
eukaryotic translation initiation factor 3 subunit E |
| chr17_-_78828893 | 0.17 |
ENST00000586066.3
|
USP36
|
ubiquitin specific peptidase 36 |
| chr1_-_158330957 | 0.17 |
ENST00000451207.5
|
CD1B
|
CD1b molecule |
| chr15_-_84716384 | 0.15 |
ENST00000559729.5
|
SEC11A
|
SEC11 homolog A, signal peptidase complex subunit |
| chr20_+_45881218 | 0.14 |
ENST00000372523.1
|
ZSWIM1
|
zinc finger SWIM-type containing 1 |
| chr12_-_84912783 | 0.14 |
ENST00000680892.1
ENST00000266682.10 ENST00000680714.1 ENST00000552192.5 |
SLC6A15
|
solute carrier family 6 member 15 |
| chr3_+_161496808 | 0.11 |
ENST00000327928.4
|
OTOL1
|
otolin 1 |
| chr15_-_84716153 | 0.09 |
ENST00000455959.7
|
SEC11A
|
SEC11 homolog A, signal peptidase complex subunit |
| chr2_+_153871909 | 0.07 |
ENST00000392825.8
ENST00000434213.1 |
GALNT13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr19_-_40714641 | 0.05 |
ENST00000678467.1
|
COQ8B
|
coenzyme Q8B |
| chr4_-_26490453 | 0.04 |
ENST00000295589.4
|
CCKAR
|
cholecystokinin A receptor |
| chr13_-_113974062 | 0.04 |
ENST00000636427.3
|
C13orf46
|
chromosome 13 open reading frame 46 |
| chr15_-_84716099 | 0.03 |
ENST00000560266.5
|
SEC11A
|
SEC11 homolog A, signal peptidase complex subunit |
| chr11_+_6259806 | 0.02 |
ENST00000532715.5
ENST00000334619.7 ENST00000525014.1 ENST00000531712.5 ENST00000525462.1 |
CCKBR
|
cholecystokinin B receptor |
| chr12_+_64780465 | 0.02 |
ENST00000542120.6
|
TBC1D30
|
TBC1 domain family member 30 |
| chr21_-_30598902 | 0.02 |
ENST00000334897.4
|
KRTAP6-2
|
keratin associated protein 6-2 |
| chr22_+_37823382 | 0.00 |
ENST00000249041.3
|
GALR3
|
galanin receptor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX20
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 29.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.9 | 3.7 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.5 | 2.8 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.3 | 1.6 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.2 | 0.6 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 0.8 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 2.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.9 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 1.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.0 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 1.4 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 4.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.9 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 3.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.4 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.2 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 1.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0015820 | leucine transport(GO:0015820) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 29.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 2.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 4.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.0 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 3.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 1.6 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 29.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 3.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 2.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 3.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) Wnt-protein binding(GO:0017147) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.0 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.1 | 1.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 4.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 1.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 3.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 3.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.4 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |