Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
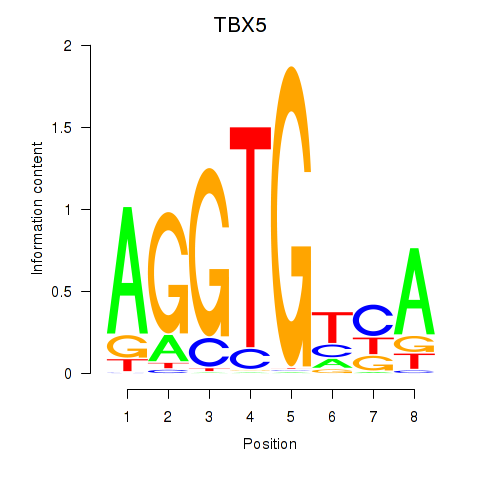
Results for TBX5
Z-value: 1.18
Transcription factors associated with TBX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX5
|
ENSG00000089225.20 | T-box transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX5 | hg38_v1_chr12_-_114406133_114406150 | 0.09 | 6.2e-01 | Click! |
Activity profile of TBX5 motif
Sorted Z-values of TBX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_45322867 | 4.37 |
ENST00000221476.4
|
CKM
|
creatine kinase, M-type |
| chr14_-_23435652 | 3.42 |
ENST00000355349.4
|
MYH7
|
myosin heavy chain 7 |
| chr11_+_1919694 | 3.39 |
ENST00000278317.11
ENST00000453458.5 ENST00000381557.6 ENST00000381579.7 ENST00000381589.7 ENST00000381563.8 ENST00000344578.8 ENST00000381558.6 |
TNNT3
|
troponin T3, fast skeletal type |
| chr11_-_107719657 | 2.98 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr12_+_80707625 | 2.97 |
ENST00000228641.4
|
MYF6
|
myogenic factor 6 |
| chr20_+_31819302 | 2.83 |
ENST00000375994.6
|
MYLK2
|
myosin light chain kinase 2 |
| chr20_+_31819348 | 2.68 |
ENST00000375985.5
|
MYLK2
|
myosin light chain kinase 2 |
| chr8_-_74321532 | 2.56 |
ENST00000342232.5
|
JPH1
|
junctophilin 1 |
| chr19_+_35031263 | 2.51 |
ENST00000640135.1
ENST00000596348.2 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr3_-_39192584 | 2.38 |
ENST00000340369.4
ENST00000421646.1 ENST00000396251.1 |
XIRP1
|
xin actin binding repeat containing 1 |
| chr11_+_1838970 | 2.03 |
ENST00000381911.6
|
TNNI2
|
troponin I2, fast skeletal type |
| chr21_+_29130630 | 2.02 |
ENST00000399926.5
ENST00000399928.6 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr21_+_29130961 | 1.97 |
ENST00000399925.5
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr22_-_50578003 | 1.90 |
ENST00000405237.7
|
CPT1B
|
carnitine palmitoyltransferase 1B |
| chr6_-_33746848 | 1.88 |
ENST00000634274.1
ENST00000293756.5 ENST00000451316.6 |
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr11_-_47449129 | 1.88 |
ENST00000298854.7
ENST00000524487.5 ENST00000529341.1 ENST00000352508.7 |
RAPSN
|
receptor associated protein of the synapse |
| chr1_+_148889403 | 1.85 |
ENST00000464103.5
ENST00000534536.5 ENST00000369356.8 ENST00000369354.7 ENST00000369347.8 ENST00000369349.7 ENST00000369351.7 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr3_-_42702638 | 1.84 |
ENST00000417472.5
ENST00000442469.1 |
HHATL
|
hedgehog acyltransferase like |
| chr17_+_39665340 | 1.83 |
ENST00000578283.1
ENST00000309889.3 |
TCAP
|
titin-cap |
| chr1_+_101236860 | 1.74 |
ENST00000475821.2
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr1_+_228208054 | 1.74 |
ENST00000284548.16
ENST00000422127.5 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr10_-_75109085 | 1.73 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chr17_+_70168610 | 1.63 |
ENST00000535240.1
|
KCNJ2
|
potassium inwardly rectifying channel subfamily J member 2 |
| chr6_+_34236865 | 1.60 |
ENST00000674029.1
ENST00000447654.5 ENST00000347617.10 ENST00000401473.7 ENST00000311487.9 |
HMGA1
|
high mobility group AT-hook 1 |
| chr1_-_16018005 | 1.58 |
ENST00000406363.2
ENST00000411503.5 ENST00000311890.14 ENST00000487046.1 |
HSPB7
|
heat shock protein family B (small) member 7 |
| chr14_-_23408265 | 1.56 |
ENST00000405093.9
|
MYH6
|
myosin heavy chain 6 |
| chr19_-_46023046 | 1.55 |
ENST00000008938.5
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr6_+_108656346 | 1.55 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr22_+_38201932 | 1.55 |
ENST00000538999.1
ENST00000538320.5 ENST00000338483.7 ENST00000441709.1 |
MAFF
|
MAF bZIP transcription factor F |
| chr16_-_2329687 | 1.53 |
ENST00000567910.1
|
ABCA3
|
ATP binding cassette subfamily A member 3 |
| chr10_+_13100075 | 1.52 |
ENST00000378747.8
ENST00000378757.6 ENST00000378752.7 ENST00000378748.7 |
OPTN
|
optineurin |
| chr13_-_113453524 | 1.48 |
ENST00000612156.2
ENST00000375418.7 |
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr7_+_80602150 | 1.46 |
ENST00000309881.11
|
CD36
|
CD36 molecule |
| chr19_-_38617928 | 1.46 |
ENST00000396857.7
ENST00000586296.5 |
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr14_-_93955577 | 1.46 |
ENST00000555507.5
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr7_+_80602200 | 1.39 |
ENST00000534394.5
|
CD36
|
CD36 molecule |
| chr4_-_185535498 | 1.39 |
ENST00000284767.12
ENST00000284771.7 ENST00000284770.10 ENST00000620787.5 ENST00000629667.2 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr1_-_153945225 | 1.38 |
ENST00000368646.6
|
DENND4B
|
DENN domain containing 4B |
| chr1_+_101237009 | 1.36 |
ENST00000305352.7
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr21_+_42219111 | 1.33 |
ENST00000450121.5
ENST00000361802.6 |
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr6_-_13487593 | 1.33 |
ENST00000379287.4
ENST00000603223.1 |
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr19_-_38617912 | 1.33 |
ENST00000591517.5
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr19_+_7677082 | 1.29 |
ENST00000597445.1
|
MCEMP1
|
mast cell expressed membrane protein 1 |
| chr19_+_7669043 | 1.27 |
ENST00000221515.6
|
RETN
|
resistin |
| chr3_+_49689531 | 1.25 |
ENST00000432042.5
ENST00000454491.5 ENST00000327697.11 |
RNF123
|
ring finger protein 123 |
| chr10_-_101229449 | 1.24 |
ENST00000370193.4
|
LBX1
|
ladybird homeobox 1 |
| chr5_-_88823763 | 1.23 |
ENST00000635898.1
ENST00000626391.2 ENST00000628656.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr17_+_70169516 | 1.22 |
ENST00000243457.4
|
KCNJ2
|
potassium inwardly rectifying channel subfamily J member 2 |
| chr22_-_50577915 | 1.21 |
ENST00000360719.6
ENST00000457250.5 |
CPT1B
|
carnitine palmitoyltransferase 1B |
| chr5_+_83471925 | 1.20 |
ENST00000502527.2
|
VCAN
|
versican |
| chr21_+_42219123 | 1.20 |
ENST00000398449.8
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr5_-_88824334 | 1.19 |
ENST00000506716.5
|
MEF2C
|
myocyte enhancer factor 2C |
| chr19_-_42217667 | 1.17 |
ENST00000336034.8
ENST00000596251.6 ENST00000598200.1 ENST00000598727.5 |
DEDD2
|
death effector domain containing 2 |
| chr1_-_206921867 | 1.15 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr5_-_100903252 | 1.15 |
ENST00000231461.10
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr19_+_50205437 | 1.15 |
ENST00000642980.1
|
MYH14
|
myosin heavy chain 14 |
| chr6_+_18387326 | 1.14 |
ENST00000259939.4
|
RNF144B
|
ring finger protein 144B |
| chr3_+_35681352 | 1.11 |
ENST00000436702.5
ENST00000438071.1 |
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr19_-_51372640 | 1.10 |
ENST00000600427.5
ENST00000221978.10 |
NKG7
|
natural killer cell granule protein 7 |
| chr22_+_38213530 | 1.07 |
ENST00000407965.1
|
MAFF
|
MAF bZIP transcription factor F |
| chr17_+_60677822 | 1.06 |
ENST00000407086.8
ENST00000589222.5 ENST00000626960.2 ENST00000390652.9 |
BCAS3
|
BCAS3 microtubule associated cell migration factor |
| chr5_-_100903214 | 1.04 |
ENST00000451528.2
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr5_+_150660841 | 1.04 |
ENST00000297130.4
|
MYOZ3
|
myozenin 3 |
| chr2_+_201129318 | 1.04 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chrY_+_12904102 | 1.02 |
ENST00000360160.8
ENST00000454054.5 |
DDX3Y
|
DEAD-box helicase 3 Y-linked |
| chr22_-_50578417 | 1.02 |
ENST00000312108.12
ENST00000395650.6 |
CPT1B
|
carnitine palmitoyltransferase 1B |
| chr2_+_29113989 | 1.01 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr1_-_16017825 | 1.01 |
ENST00000463576.5
|
HSPB7
|
heat shock protein family B (small) member 7 |
| chr2_-_85668172 | 1.01 |
ENST00000428225.5
ENST00000519937.7 |
SFTPB
|
surfactant protein B |
| chr21_+_15842393 | 1.01 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chrX_+_96684712 | 1.01 |
ENST00000373049.8
|
DIAPH2
|
diaphanous related formin 2 |
| chrX_+_96684638 | 1.01 |
ENST00000355827.8
ENST00000373061.7 |
DIAPH2
|
diaphanous related formin 2 |
| chr19_-_39833615 | 1.01 |
ENST00000593685.5
ENST00000600611.5 |
DYRK1B
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr5_-_143405323 | 1.00 |
ENST00000514699.1
|
NR3C1
|
nuclear receptor subfamily 3 group C member 1 |
| chr11_+_46332905 | 1.00 |
ENST00000343674.10
|
DGKZ
|
diacylglycerol kinase zeta |
| chr11_+_35176575 | 0.99 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr3_+_42653690 | 0.99 |
ENST00000232974.11
|
ZBTB47
|
zinc finger and BTB domain containing 47 |
| chr14_+_75280078 | 0.99 |
ENST00000555347.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chrX_+_96684929 | 0.98 |
ENST00000373054.5
|
DIAPH2
|
diaphanous related formin 2 |
| chr19_-_49441508 | 0.98 |
ENST00000221485.8
|
SLC17A7
|
solute carrier family 17 member 7 |
| chr3_-_187745460 | 0.98 |
ENST00000406870.7
|
BCL6
|
BCL6 transcription repressor |
| chr10_-_75073627 | 0.97 |
ENST00000338487.6
|
DUSP29
|
dual specificity phosphatase 29 |
| chr11_+_35176696 | 0.96 |
ENST00000528455.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_-_150010675 | 0.95 |
ENST00000417191.2
ENST00000581312.6 |
OTUD7B
|
OTU deubiquitinase 7B |
| chr3_+_193241190 | 0.93 |
ENST00000264735.4
|
PLAAT1
|
phospholipase A and acyltransferase 1 |
| chr6_-_46015812 | 0.93 |
ENST00000544153.3
ENST00000339561.12 |
CLIC5
|
chloride intracellular channel 5 |
| chr11_+_35176611 | 0.92 |
ENST00000279452.10
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_-_6209389 | 0.92 |
ENST00000465335.1
|
RPL22
|
ribosomal protein L22 |
| chr12_-_116276759 | 0.92 |
ENST00000548743.2
|
MED13L
|
mediator complex subunit 13L |
| chr19_-_51372686 | 0.92 |
ENST00000595217.1
|
NKG7
|
natural killer cell granule protein 7 |
| chr18_+_61333424 | 0.91 |
ENST00000262717.9
|
CDH20
|
cadherin 20 |
| chr20_+_836052 | 0.91 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110 member A |
| chr11_+_35176639 | 0.91 |
ENST00000527889.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr6_-_41201128 | 0.90 |
ENST00000483722.2
|
TREML2
|
triggering receptor expressed on myeloid cells like 2 |
| chr3_+_193241128 | 0.90 |
ENST00000650797.1
|
PLAAT1
|
phospholipase A and acyltransferase 1 |
| chr15_+_88638947 | 0.90 |
ENST00000559876.2
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chrX_+_21374357 | 0.89 |
ENST00000643841.1
ENST00000379510.5 ENST00000425654.7 ENST00000644798.1 ENST00000543067.6 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr1_+_202462730 | 0.89 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr19_+_7669080 | 0.89 |
ENST00000629642.1
|
RETN
|
resistin |
| chr12_+_109131350 | 0.89 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chrX_+_96684801 | 0.88 |
ENST00000324765.13
|
DIAPH2
|
diaphanous related formin 2 |
| chr4_-_101347551 | 0.88 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr6_-_46015607 | 0.87 |
ENST00000644878.1
ENST00000644324.1 ENST00000672327.1 |
CLIC5
|
chloride intracellular channel 5 |
| chr5_+_139795795 | 0.86 |
ENST00000274710.4
|
PSD2
|
pleckstrin and Sec7 domain containing 2 |
| chr15_+_88639009 | 0.85 |
ENST00000306072.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr19_-_42427379 | 0.84 |
ENST00000244289.9
|
LIPE
|
lipase E, hormone sensitive type |
| chr9_+_126335200 | 0.83 |
ENST00000402437.2
|
MVB12B
|
multivesicular body subunit 12B |
| chr2_+_173354820 | 0.82 |
ENST00000347703.7
ENST00000410101.7 ENST00000410019.3 ENST00000306721.8 |
CDCA7
|
cell division cycle associated 7 |
| chr17_+_27471999 | 0.82 |
ENST00000583370.5
ENST00000509603.6 ENST00000268763.10 ENST00000398988.7 |
KSR1
|
kinase suppressor of ras 1 |
| chr5_-_88824266 | 0.81 |
ENST00000509373.1
ENST00000636541.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr1_-_32702736 | 0.81 |
ENST00000373484.4
ENST00000409190.8 |
SYNC
|
syncoilin, intermediate filament protein |
| chrX_+_21374608 | 0.80 |
ENST00000644295.1
ENST00000645074.1 ENST00000645791.1 ENST00000643220.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr11_+_73647549 | 0.79 |
ENST00000227214.10
ENST00000398494.8 ENST00000543085.5 |
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr1_-_10982037 | 0.79 |
ENST00000377004.8
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr10_-_31031911 | 0.79 |
ENST00000375311.1
ENST00000413025.5 ENST00000436087.6 ENST00000452305.5 ENST00000442986.5 |
ZNF438
|
zinc finger protein 438 |
| chr20_+_59300402 | 0.78 |
ENST00000311585.11
ENST00000371028.6 |
EDN3
|
endothelin 3 |
| chr8_+_122781621 | 0.78 |
ENST00000314393.6
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr1_+_203026481 | 0.78 |
ENST00000367240.6
|
PPFIA4
|
PTPRF interacting protein alpha 4 |
| chr8_+_127737610 | 0.77 |
ENST00000652288.1
|
MYC
|
MYC proto-oncogene, bHLH transcription factor |
| chr7_+_74183814 | 0.77 |
ENST00000678341.1
|
EIF4H
|
eukaryotic translation initiation factor 4H |
| chr3_+_167735238 | 0.77 |
ENST00000472941.5
|
SERPINI1
|
serpin family I member 1 |
| chr1_-_151146643 | 0.77 |
ENST00000613223.1
|
SEMA6C
|
semaphorin 6C |
| chr1_-_151146611 | 0.75 |
ENST00000341697.7
ENST00000368914.8 |
SEMA6C
|
semaphorin 6C |
| chr7_+_143316105 | 0.75 |
ENST00000343257.7
ENST00000650516.1 |
CLCN1
|
chloride voltage-gated channel 1 |
| chr15_+_90872162 | 0.75 |
ENST00000680053.1
|
FURIN
|
furin, paired basic amino acid cleaving enzyme |
| chr3_-_18424533 | 0.75 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr22_-_38754941 | 0.74 |
ENST00000420859.5
ENST00000452294.5 ENST00000456894.5 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr17_-_7329266 | 0.74 |
ENST00000571887.5
ENST00000315614.11 ENST00000399464.7 ENST00000570460.5 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr5_+_83471668 | 0.73 |
ENST00000342785.8
ENST00000343200.9 |
VCAN
|
versican |
| chr5_-_88884525 | 0.73 |
ENST00000502983.5
|
MEF2C
|
myocyte enhancer factor 2C |
| chr20_+_59300589 | 0.71 |
ENST00000337938.7
ENST00000371025.7 |
EDN3
|
endothelin 3 |
| chr3_+_134795277 | 0.71 |
ENST00000647596.1
|
EPHB1
|
EPH receptor B1 |
| chr20_+_58888779 | 0.70 |
ENST00000488546.6
ENST00000667293.2 ENST00000481039.6 ENST00000467321.6 |
GNAS
|
GNAS complex locus |
| chr17_-_1684807 | 0.70 |
ENST00000577001.1
ENST00000572621.5 ENST00000304992.11 |
PRPF8
|
pre-mRNA processing factor 8 |
| chr6_-_31652115 | 0.70 |
ENST00000454165.1
ENST00000428326.5 ENST00000452994.5 |
BAG6
|
BAG cochaperone 6 |
| chr14_+_90398159 | 0.69 |
ENST00000544280.6
|
CALM1
|
calmodulin 1 |
| chr2_+_120346130 | 0.69 |
ENST00000295228.4
|
INHBB
|
inhibin subunit beta B |
| chr18_+_34493289 | 0.69 |
ENST00000682923.1
ENST00000596745.5 ENST00000283365.14 ENST00000315456.10 ENST00000598774.6 ENST00000684266.1 ENST00000683092.1 ENST00000683379.1 ENST00000684359.1 |
DTNA
|
dystrobrevin alpha |
| chrX_-_107775740 | 0.69 |
ENST00000372383.9
|
TSC22D3
|
TSC22 domain family member 3 |
| chr7_+_16661182 | 0.68 |
ENST00000446596.5
ENST00000452975.6 ENST00000438834.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr14_+_71932397 | 0.68 |
ENST00000553525.6
ENST00000555571.5 |
RGS6
|
regulator of G protein signaling 6 |
| chr6_-_31651964 | 0.68 |
ENST00000433828.5
ENST00000456286.5 |
BAG6
|
BAG cochaperone 6 |
| chrX_-_107717054 | 0.67 |
ENST00000503515.1
ENST00000372397.6 |
TSC22D3
|
TSC22 domain family member 3 |
| chr6_-_31651920 | 0.67 |
ENST00000434444.5
|
BAG6
|
BAG cochaperone 6 |
| chr19_+_35138993 | 0.67 |
ENST00000612146.4
ENST00000589209.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr3_+_134795248 | 0.66 |
ENST00000398015.8
|
EPHB1
|
EPH receptor B1 |
| chr19_+_49877660 | 0.66 |
ENST00000535102.6
|
TBC1D17
|
TBC1 domain family member 17 |
| chr2_+_56183973 | 0.66 |
ENST00000407595.3
|
CCDC85A
|
coiled-coil domain containing 85A |
| chr19_+_49877425 | 0.66 |
ENST00000622860.4
|
TBC1D17
|
TBC1 domain family member 17 |
| chrX_-_107716401 | 0.65 |
ENST00000486554.1
ENST00000372390.8 |
TSC22D3
|
TSC22 domain family member 3 |
| chr22_-_30266839 | 0.65 |
ENST00000403463.1
ENST00000215781.3 |
OSM
|
oncostatin M |
| chr12_-_52493250 | 0.65 |
ENST00000330722.7
|
KRT6A
|
keratin 6A |
| chr16_+_30664334 | 0.65 |
ENST00000287468.5
|
FBRS
|
fibrosin |
| chr19_-_47231191 | 0.64 |
ENST00000439096.3
|
BBC3
|
BCL2 binding component 3 |
| chr16_+_2496032 | 0.64 |
ENST00000564543.1
|
ENSG00000260272.1
|
novel protein |
| chr11_-_123654581 | 0.64 |
ENST00000392770.6
ENST00000530277.5 ENST00000299333.8 |
SCN3B
|
sodium voltage-gated channel beta subunit 3 |
| chr7_-_82443715 | 0.64 |
ENST00000356253.9
ENST00000423588.1 |
CACNA2D1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr19_-_42132391 | 0.64 |
ENST00000528894.8
ENST00000560804.6 ENST00000560558.5 ENST00000560398.5 ENST00000526816.6 ENST00000625670.2 |
POU2F2
|
POU class 2 homeobox 2 |
| chr5_+_150661243 | 0.64 |
ENST00000517768.6
|
MYOZ3
|
myozenin 3 |
| chr7_+_142500027 | 0.63 |
ENST00000390375.2
|
TRBV5-6
|
T cell receptor beta variable 5-6 |
| chr17_+_80545422 | 0.63 |
ENST00000544334.6
|
RPTOR
|
regulatory associated protein of MTOR complex 1 |
| chr17_+_80544817 | 0.63 |
ENST00000306801.8
ENST00000570891.5 |
RPTOR
|
regulatory associated protein of MTOR complex 1 |
| chr16_+_2969307 | 0.62 |
ENST00000576565.1
ENST00000318782.9 |
PAQR4
|
progestin and adipoQ receptor family member 4 |
| chr12_-_120225989 | 0.62 |
ENST00000546532.5
ENST00000548912.5 |
PXN
|
paxillin |
| chr19_-_38899710 | 0.62 |
ENST00000447739.1
ENST00000407552.5 |
SIRT2
|
sirtuin 2 |
| chr6_-_31592952 | 0.61 |
ENST00000376073.8
ENST00000376072.7 |
NCR3
|
natural cytotoxicity triggering receptor 3 |
| chr5_+_55024250 | 0.61 |
ENST00000231009.3
|
GZMK
|
granzyme K |
| chr17_-_35089212 | 0.61 |
ENST00000584655.5
ENST00000447669.6 ENST00000315249.11 |
RFFL
|
ring finger and FYVE like domain containing E3 ubiquitin protein ligase |
| chr2_+_201129483 | 0.61 |
ENST00000440180.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr20_+_59300547 | 0.60 |
ENST00000644821.1
|
EDN3
|
endothelin 3 |
| chr3_+_69739425 | 0.59 |
ENST00000352241.9
ENST00000642352.1 |
MITF
|
melanocyte inducing transcription factor |
| chr7_+_142592928 | 0.59 |
ENST00000616518.1
|
TRBV15
|
T cell receptor beta variable 15 |
| chr2_+_68734861 | 0.58 |
ENST00000467265.5
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr4_-_101347492 | 0.58 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr6_-_31592992 | 0.58 |
ENST00000340027.10
|
NCR3
|
natural cytotoxicity triggering receptor 3 |
| chrX_-_50470818 | 0.58 |
ENST00000611977.2
|
DGKK
|
diacylglycerol kinase kappa |
| chr5_+_161850597 | 0.57 |
ENST00000634335.1
ENST00000635880.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr11_-_41459592 | 0.57 |
ENST00000528697.6
ENST00000530763.5 |
LRRC4C
|
leucine rich repeat containing 4C |
| chr14_+_22281097 | 0.57 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chrX_-_107775951 | 0.56 |
ENST00000315660.8
ENST00000372384.6 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family member 3 |
| chrX_+_21374476 | 0.56 |
ENST00000644585.1
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr1_-_205321737 | 0.56 |
ENST00000367157.6
|
NUAK2
|
NUAK family kinase 2 |
| chr7_-_82443766 | 0.56 |
ENST00000356860.8
|
CACNA2D1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr17_+_57105899 | 0.56 |
ENST00000576295.5
|
AKAP1
|
A-kinase anchoring protein 1 |
| chr4_-_101347327 | 0.55 |
ENST00000394853.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chrX_-_55030970 | 0.55 |
ENST00000493869.2
ENST00000396198.7 ENST00000650242.1 ENST00000335854.8 ENST00000477869.6 ENST00000455688.2 ENST00000644983.1 |
ALAS2
|
5'-aminolevulinate synthase 2 |
| chr21_+_42514523 | 0.55 |
ENST00000398343.2
|
SLC37A1
|
solute carrier family 37 member 1 |
| chr11_+_119107335 | 0.55 |
ENST00000648610.2
ENST00000336702.7 |
C2CD2L
|
C2CD2 like |
| chr16_-_50681206 | 0.54 |
ENST00000610485.1
|
SNX20
|
sorting nexin 20 |
| chr19_-_48364034 | 0.54 |
ENST00000435956.7
|
TMEM143
|
transmembrane protein 143 |
| chr1_-_16352420 | 0.54 |
ENST00000375592.8
|
FBXO42
|
F-box protein 42 |
| chr7_-_6026552 | 0.54 |
ENST00000422786.1
|
EIF2AK1
|
eukaryotic translation initiation factor 2 alpha kinase 1 |
| chr15_-_70763430 | 0.53 |
ENST00000539319.5
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr16_-_50681289 | 0.53 |
ENST00000423026.6
ENST00000330943.9 |
SNX20
|
sorting nexin 20 |
| chr15_+_81296913 | 0.53 |
ENST00000394652.6
|
IL16
|
interleukin 16 |
| chr18_+_34493904 | 0.53 |
ENST00000680366.1
ENST00000684560.1 |
DTNA
|
dystrobrevin alpha |
| chr1_-_153545793 | 0.53 |
ENST00000354332.8
ENST00000368716.9 |
S100A4
|
S100 calcium binding protein A4 |
| chr1_+_111619751 | 0.53 |
ENST00000433097.5
ENST00000369709.3 |
RAP1A
|
RAP1A, member of RAS oncogene family |
| chr15_+_90872862 | 0.53 |
ENST00000618099.4
|
FURIN
|
furin, paired basic amino acid cleaving enzyme |
| chr4_+_94489030 | 0.52 |
ENST00000510099.5
|
PDLIM5
|
PDZ and LIM domain 5 |
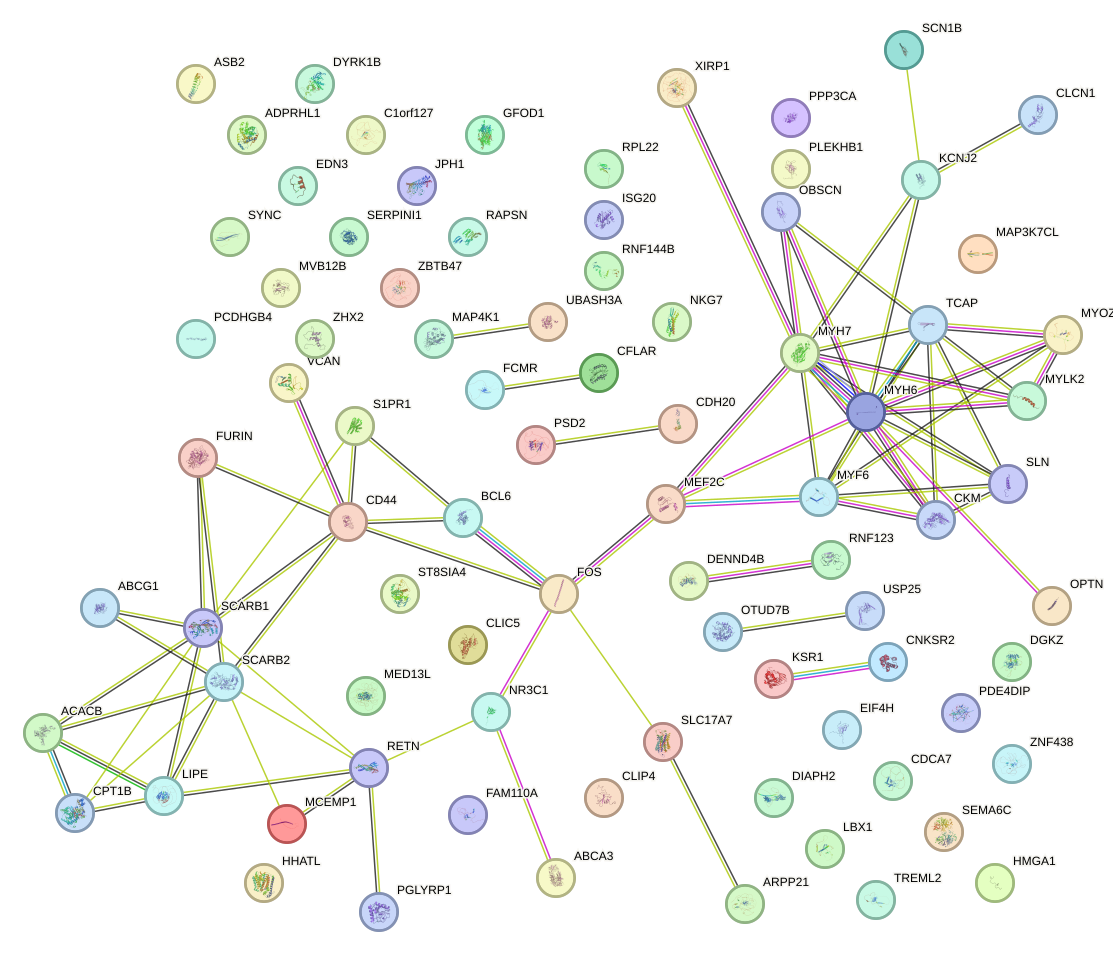
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 2.9 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 1.0 | 3.0 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.9 | 3.4 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.8 | 2.5 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.8 | 5.5 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.6 | 3.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.6 | 2.2 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.5 | 2.2 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.5 | 1.5 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.5 | 0.5 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.5 | 2.5 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.5 | 1.8 | GO:0060262 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.5 | 1.4 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.4 | 1.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 2.5 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.4 | 1.1 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.4 | 1.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.4 | 4.0 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.3 | 1.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.3 | 1.6 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.3 | 1.3 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.3 | 2.6 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 3.8 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.3 | 2.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.3 | 0.8 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.3 | 4.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 0.7 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.2 | 1.8 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 2.9 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 0.7 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.2 | 2.9 | GO:2000332 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 0.4 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.2 | 1.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 1.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.2 | 1.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.2 | 1.0 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.2 | 0.8 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.2 | 1.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 2.0 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.2 | 1.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 1.2 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.2 | 1.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 3.8 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 1.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.8 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.5 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 1.7 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.1 | 0.4 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.1 | 1.6 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.1 | 0.5 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 1.6 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 1.3 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.1 | 0.9 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.3 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.1 | 0.9 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 0.8 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.2 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 2.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.3 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.1 | 0.3 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.1 | 5.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.7 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 6.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.6 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.3 | GO:0021722 | pons maturation(GO:0021586) superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.2 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.1 | 2.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 1.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.5 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 2.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.5 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.7 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 1.9 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.5 | GO:0097327 | response to antineoplastic agent(GO:0097327) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.4 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.5 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.3 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.8 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.1 | 0.3 | GO:0002554 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.1 | 0.3 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 0.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 1.0 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.1 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 0.2 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.1 | 0.3 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.4 | GO:0045759 | negative regulation of action potential(GO:0045759) negative regulation of transmission of nerve impulse(GO:0051970) regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.4 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 0.8 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.2 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 1.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.3 | GO:0046778 | modification by virus of host mRNA processing(GO:0046778) |
| 0.1 | 0.8 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 2.6 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 0.3 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.5 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 1.0 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.5 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.3 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.2 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 | 1.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.6 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 2.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.8 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 4.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.4 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.7 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.6 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.3 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 2.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.2 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 3.3 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.7 | GO:0048757 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.5 | GO:0070262 | mitotic nuclear envelope reassembly(GO:0007084) peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.8 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.4 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0090237 | regulation of arachidonic acid secretion(GO:0090237) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.1 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.6 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.0 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.1 | GO:0060161 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 1.7 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 1.8 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.5 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.4 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.6 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.7 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.5 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.5 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.0 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 1.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 1.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.3 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 1.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 1.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.0 | GO:1902938 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.2 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.9 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.4 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.1 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.4 | 1.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.3 | 3.8 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 1.6 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.3 | 5.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 6.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 2.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 2.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 2.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 1.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.5 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.4 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 1.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 1.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 3.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 1.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 2.0 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.3 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 1.0 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.1 | 0.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.2 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 1.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.5 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 2.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 4.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 3.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 1.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 1.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.7 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 14.8 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 2.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 3.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.9 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 5.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 4.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 1.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.5 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 2.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 2.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 2.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 1.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.6 | 1.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.6 | 3.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.6 | 5.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 4.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 4.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.5 | 2.5 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.4 | 3.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.4 | 1.2 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.4 | 1.6 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.4 | 1.1 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.4 | 1.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.3 | 2.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 1.7 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.3 | 1.3 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.3 | 2.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 0.8 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.3 | 0.8 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.3 | 1.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.2 | 1.5 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.2 | 2.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 5.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 2.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 0.7 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 6.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 0.9 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.2 | 0.6 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.2 | 2.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 3.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 2.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 1.0 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.9 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 1.0 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 2.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 2.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.6 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 1.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.7 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 2.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.3 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.3 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 4.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 3.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 1.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.6 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 1.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 1.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 2.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 5.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.5 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.8 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 1.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 2.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.8 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 1.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.0 | 0.3 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 3.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0044020 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0031811 | beta-1 adrenergic receptor binding(GO:0031697) G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0034594 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 1.0 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 3.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 9.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 4.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 2.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 5.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 2.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 2.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 2.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 4.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 2.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 5.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 4.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 2.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 2.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 1.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 2.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 3.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 2.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 3.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 2.7 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.7 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 2.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |