Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
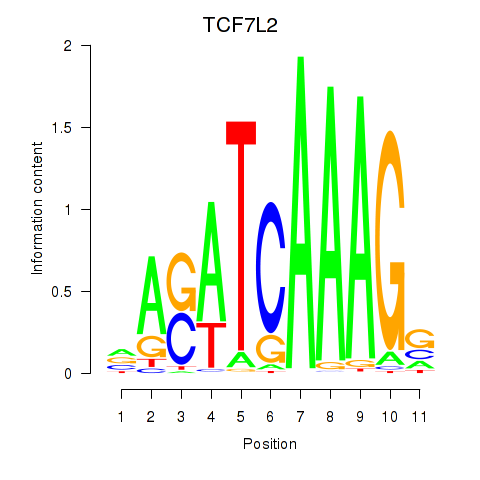
Results for TCF7L2
Z-value: 0.90
Transcription factors associated with TCF7L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L2
|
ENSG00000148737.17 | transcription factor 7 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L2 | hg38_v1_chr10_+_113125536_113125665 | -0.24 | 1.8e-01 | Click! |
Activity profile of TCF7L2 motif
Sorted Z-values of TCF7L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_72063226 | 4.69 |
ENST00000540303.7
ENST00000356967.6 ENST00000561690.1 |
HPR
|
haptoglobin-related protein |
| chr11_+_114257800 | 4.53 |
ENST00000535401.5
|
NNMT
|
nicotinamide N-methyltransferase |
| chr11_+_69641146 | 3.92 |
ENST00000227507.3
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr16_+_72054477 | 3.55 |
ENST00000355906.10
ENST00000570083.5 ENST00000228226.12 ENST00000398131.6 ENST00000569639.5 ENST00000564499.5 ENST00000357763.8 ENST00000613898.1 ENST00000562526.5 ENST00000565574.5 ENST00000568417.6 |
HP
|
haptoglobin |
| chr6_+_31946086 | 3.28 |
ENST00000425368.7
|
CFB
|
complement factor B |
| chr2_-_88128049 | 3.17 |
ENST00000393750.3
ENST00000295834.8 |
FABP1
|
fatty acid binding protein 1 |
| chr10_+_7703300 | 3.13 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_113553039 | 3.07 |
ENST00000351270.4
|
HABP2
|
hyaluronan binding protein 2 |
| chrX_-_32155462 | 3.01 |
ENST00000359836.5
ENST00000378707.7 ENST00000541735.5 ENST00000684130.1 ENST00000682238.1 ENST00000620040.5 ENST00000474231.5 |
DMD
|
dystrophin |
| chr2_+_233712905 | 2.99 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr10_+_7703340 | 2.90 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr5_-_135954962 | 2.75 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr17_-_69141878 | 2.51 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr14_-_64942783 | 2.23 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2 |
| chr17_+_1771688 | 2.13 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin family F member 1 |
| chr7_+_138076453 | 2.05 |
ENST00000242375.8
ENST00000411726.6 |
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chrX_+_153494970 | 2.02 |
ENST00000331595.9
ENST00000431891.1 |
BGN
|
biglycan |
| chrX_+_47078380 | 1.97 |
ENST00000352078.8
|
RGN
|
regucalcin |
| chrX_+_47078330 | 1.94 |
ENST00000457380.5
|
RGN
|
regucalcin |
| chr1_+_99646025 | 1.94 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chrX_+_47078434 | 1.86 |
ENST00000397180.6
|
RGN
|
regucalcin |
| chr4_-_108762964 | 1.84 |
ENST00000512646.5
ENST00000411864.6 ENST00000296486.8 ENST00000510706.5 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr5_-_154478218 | 1.81 |
ENST00000231121.3
|
HAND1
|
heart and neural crest derivatives expressed 1 |
| chr12_+_107774704 | 1.72 |
ENST00000342331.5
|
ASCL4
|
achaete-scute family bHLH transcription factor 4 |
| chr7_+_138076422 | 1.52 |
ENST00000432161.5
|
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chr10_+_99782628 | 1.46 |
ENST00000648689.1
ENST00000647814.1 |
ABCC2
|
ATP binding cassette subfamily C member 2 |
| chr13_+_75760659 | 1.43 |
ENST00000526202.5
ENST00000465261.6 |
LMO7
|
LIM domain 7 |
| chr17_-_65561137 | 1.42 |
ENST00000580513.1
|
AXIN2
|
axin 2 |
| chr2_+_167135901 | 1.41 |
ENST00000628543.2
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr4_-_74099187 | 1.39 |
ENST00000508487.3
|
CXCL2
|
C-X-C motif chemokine ligand 2 |
| chr14_-_93976719 | 1.34 |
ENST00000555287.1
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr17_+_44004604 | 1.31 |
ENST00000293404.8
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr17_+_81712236 | 1.29 |
ENST00000545862.5
ENST00000350690.10 ENST00000331531.9 |
SLC25A10
|
solute carrier family 25 member 10 |
| chr7_-_149028452 | 1.28 |
ENST00000413966.1
ENST00000652332.1 |
PDIA4
|
protein disulfide isomerase family A member 4 |
| chr13_+_75760431 | 1.27 |
ENST00000321797.12
|
LMO7
|
LIM domain 7 |
| chr10_-_86957582 | 1.25 |
ENST00000372027.10
|
MMRN2
|
multimerin 2 |
| chr1_+_162381703 | 1.22 |
ENST00000458626.4
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr13_-_27969295 | 1.20 |
ENST00000381020.8
|
CDX2
|
caudal type homeobox 2 |
| chr6_-_25930678 | 1.19 |
ENST00000377850.8
|
SLC17A2
|
solute carrier family 17 member 2 |
| chr7_-_149028651 | 1.18 |
ENST00000286091.9
|
PDIA4
|
protein disulfide isomerase family A member 4 |
| chr6_-_25930611 | 1.16 |
ENST00000360488.7
|
SLC17A2
|
solute carrier family 17 member 2 |
| chrX_+_106920393 | 1.15 |
ENST00000336803.2
|
CLDN2
|
claudin 2 |
| chr18_+_48539017 | 1.14 |
ENST00000256413.8
|
CTIF
|
cap binding complex dependent translation initiation factor |
| chr8_-_118951876 | 1.07 |
ENST00000297350.9
|
TNFRSF11B
|
TNF receptor superfamily member 11b |
| chr9_-_4741176 | 1.06 |
ENST00000381809.8
|
AK3
|
adenylate kinase 3 |
| chr2_-_71130214 | 1.04 |
ENST00000494660.6
ENST00000244217.6 ENST00000486135.1 |
MCEE
|
methylmalonyl-CoA epimerase |
| chr17_-_47957824 | 1.03 |
ENST00000300557.3
|
PRR15L
|
proline rich 15 like |
| chr4_-_64409444 | 1.02 |
ENST00000381210.8
ENST00000507440.5 |
TECRL
|
trans-2,3-enoyl-CoA reductase like |
| chr16_-_73048104 | 1.02 |
ENST00000268489.10
|
ZFHX3
|
zinc finger homeobox 3 |
| chr8_-_141002072 | 0.99 |
ENST00000517453.5
|
PTK2
|
protein tyrosine kinase 2 |
| chr14_-_93976550 | 0.95 |
ENST00000555019.6
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr7_+_94394886 | 0.92 |
ENST00000297268.11
ENST00000620463.1 |
COL1A2
|
collagen type I alpha 2 chain |
| chr3_+_29281552 | 0.89 |
ENST00000452462.5
ENST00000456853.1 |
RBMS3
|
RNA binding motif single stranded interacting protein 3 |
| chr5_-_176388563 | 0.89 |
ENST00000509257.1
ENST00000616685.1 ENST00000614830.5 |
NOP16
|
NOP16 nucleolar protein |
| chr15_-_89751292 | 0.88 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior bHLH transcription factor 1 |
| chr1_+_60865259 | 0.87 |
ENST00000371191.5
|
NFIA
|
nuclear factor I A |
| chr20_-_57265738 | 0.87 |
ENST00000433911.1
|
BMP7
|
bone morphogenetic protein 7 |
| chr2_+_69013170 | 0.86 |
ENST00000303714.9
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr18_+_48539198 | 0.85 |
ENST00000591412.5
|
CTIF
|
cap binding complex dependent translation initiation factor |
| chr18_-_26865689 | 0.85 |
ENST00000675739.1
ENST00000383168.9 ENST00000672981.2 ENST00000578776.1 |
AQP4
|
aquaporin 4 |
| chr4_-_64409381 | 0.85 |
ENST00000509536.1
|
TECRL
|
trans-2,3-enoyl-CoA reductase like |
| chr1_-_109392893 | 0.83 |
ENST00000633956.1
|
SORT1
|
sortilin 1 |
| chr14_+_32329341 | 0.83 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr5_-_59039454 | 0.80 |
ENST00000358923.10
|
PDE4D
|
phosphodiesterase 4D |
| chr18_+_48539112 | 0.79 |
ENST00000382998.8
|
CTIF
|
cap binding complex dependent translation initiation factor |
| chr9_+_125748175 | 0.78 |
ENST00000491787.7
ENST00000447726.6 |
PBX3
|
PBX homeobox 3 |
| chr19_+_49581304 | 0.77 |
ENST00000246794.10
|
PRRG2
|
proline rich and Gla domain 2 |
| chr4_+_86936386 | 0.76 |
ENST00000511442.1
|
AFF1
|
AF4/FMR2 family member 1 |
| chr16_+_19211157 | 0.75 |
ENST00000568433.1
|
SYT17
|
synaptotagmin 17 |
| chr5_-_176388629 | 0.75 |
ENST00000619979.4
ENST00000621444.4 |
NOP16
|
NOP16 nucleolar protein |
| chr2_+_112911159 | 0.75 |
ENST00000263326.8
|
IL37
|
interleukin 37 |
| chr14_+_32329256 | 0.74 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr10_-_73591330 | 0.74 |
ENST00000451492.5
ENST00000681793.1 ENST00000680396.1 ENST00000413442.5 |
USP54
|
ubiquitin specific peptidase 54 |
| chr19_+_54983486 | 0.74 |
ENST00000540005.1
|
NLRP2
|
NLR family pyrin domain containing 2 |
| chr6_+_63571702 | 0.73 |
ENST00000672924.1
|
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr7_+_138076475 | 0.73 |
ENST00000438242.1
|
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chr1_+_77918128 | 0.72 |
ENST00000342754.5
|
NEXN
|
nexilin F-actin binding protein |
| chr2_+_69013414 | 0.71 |
ENST00000681816.1
ENST00000482235.2 |
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr1_-_11847772 | 0.70 |
ENST00000376480.7
ENST00000610706.1 |
NPPA
|
natriuretic peptide A |
| chr7_+_120988683 | 0.69 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr18_-_26865732 | 0.69 |
ENST00000672188.1
|
AQP4
|
aquaporin 4 |
| chr10_-_102432565 | 0.68 |
ENST00000369937.5
|
CUEDC2
|
CUE domain containing 2 |
| chrX_-_47145035 | 0.68 |
ENST00000276062.8
|
NDUFB11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chrX_-_47144680 | 0.67 |
ENST00000377811.4
|
NDUFB11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr14_-_52791597 | 0.66 |
ENST00000216410.8
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr5_+_171419635 | 0.66 |
ENST00000274625.6
|
FGF18
|
fibroblast growth factor 18 |
| chr15_+_43593601 | 0.65 |
ENST00000449946.5
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr10_+_52128343 | 0.65 |
ENST00000672084.1
|
PRKG1
|
protein kinase cGMP-dependent 1 |
| chr15_-_55588337 | 0.65 |
ENST00000563719.4
|
PYGO1
|
pygopus family PHD finger 1 |
| chr2_-_55917699 | 0.65 |
ENST00000634374.1
|
EFEMP1
|
EGF containing fibulin extracellular matrix protein 1 |
| chr10_+_112374110 | 0.62 |
ENST00000354655.9
|
ACSL5
|
acyl-CoA synthetase long chain family member 5 |
| chr3_-_134374439 | 0.60 |
ENST00000513145.1
ENST00000249883.10 ENST00000422605.6 |
AMOTL2
|
angiomotin like 2 |
| chr15_+_43593054 | 0.60 |
ENST00000453782.5
ENST00000300283.10 ENST00000437924.5 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr2_+_69013379 | 0.59 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr19_-_40285251 | 0.58 |
ENST00000358335.9
|
AKT2
|
AKT serine/threonine kinase 2 |
| chr2_+_170715317 | 0.58 |
ENST00000375281.4
|
SP5
|
Sp5 transcription factor |
| chr10_-_99620401 | 0.56 |
ENST00000370495.6
|
SLC25A28
|
solute carrier family 25 member 28 |
| chr14_-_52791462 | 0.56 |
ENST00000650397.1
ENST00000554230.5 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr1_+_154272589 | 0.56 |
ENST00000457918.6
ENST00000483970.6 ENST00000328703.12 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr17_-_81961837 | 0.56 |
ENST00000425009.1
|
NOTUM
|
notum, palmitoleoyl-protein carboxylesterase |
| chr1_-_93614091 | 0.56 |
ENST00000370247.7
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr1_-_11848345 | 0.55 |
ENST00000376476.1
|
NPPA
|
natriuretic peptide A |
| chr19_-_40285277 | 0.54 |
ENST00000579047.5
ENST00000392038.7 |
AKT2
|
AKT serine/threonine kinase 2 |
| chr6_-_11779606 | 0.52 |
ENST00000506810.1
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chrX_+_2752024 | 0.52 |
ENST00000644266.2
ENST00000419513.7 ENST00000509484.3 ENST00000381174.10 |
XG
|
Xg glycoprotein (Xg blood group) |
| chr20_-_675793 | 0.51 |
ENST00000488788.2
ENST00000246104.7 |
ENSG00000270299.1
SCRT2
|
novel protein scratch family transcriptional repressor 2 |
| chr4_+_30720348 | 0.51 |
ENST00000361762.3
|
PCDH7
|
protocadherin 7 |
| chr5_-_16936231 | 0.51 |
ENST00000507288.1
ENST00000274203.13 ENST00000513610.6 |
MYO10
|
myosin X |
| chr2_+_69013337 | 0.51 |
ENST00000463335.2
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr11_-_108552196 | 0.50 |
ENST00000526312.5
|
EXPH5
|
exophilin 5 |
| chr19_-_18606779 | 0.50 |
ENST00000684169.1
ENST00000392386.8 |
CRLF1
|
cytokine receptor like factor 1 |
| chr5_-_149551381 | 0.48 |
ENST00000670598.1
ENST00000657001.1 ENST00000515768.6 ENST00000261798.10 ENST00000377843.8 |
CSNK1A1
|
casein kinase 1 alpha 1 |
| chr3_+_217535 | 0.48 |
ENST00000449294.6
|
CHL1
|
cell adhesion molecule L1 like |
| chrX_+_17656082 | 0.48 |
ENST00000648929.1
|
NHS
|
NHS actin remodeling regulator |
| chr19_-_40285395 | 0.48 |
ENST00000424901.5
ENST00000578123.5 |
AKT2
|
AKT serine/threonine kinase 2 |
| chr19_-_43504711 | 0.47 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology like domain family B member 3 |
| chrX_+_111511661 | 0.47 |
ENST00000569275.1
|
SERTM2
|
serine rich and transmembrane domain containing 2 |
| chr11_-_70717994 | 0.47 |
ENST00000659264.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr14_+_32934383 | 0.47 |
ENST00000551634.6
|
NPAS3
|
neuronal PAS domain protein 3 |
| chr22_+_40951364 | 0.47 |
ENST00000216225.9
|
RBX1
|
ring-box 1 |
| chr11_+_111878926 | 0.46 |
ENST00000528125.5
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr15_+_92904447 | 0.46 |
ENST00000626782.2
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr8_-_6926066 | 0.46 |
ENST00000297436.3
|
DEFA6
|
defensin alpha 6 |
| chr7_+_80646305 | 0.43 |
ENST00000426978.5
ENST00000432207.5 |
CD36
|
CD36 molecule |
| chr5_+_102865737 | 0.42 |
ENST00000509523.2
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr4_+_112860912 | 0.41 |
ENST00000671951.1
|
ANK2
|
ankyrin 2 |
| chr3_-_155293665 | 0.41 |
ENST00000489090.2
|
STRIT1
|
small transmembrane regulator of ion transport 1 |
| chr17_-_41350824 | 0.41 |
ENST00000007735.4
|
KRT33A
|
keratin 33A |
| chr4_+_112861053 | 0.41 |
ENST00000672221.1
|
ANK2
|
ankyrin 2 |
| chr20_+_59300547 | 0.40 |
ENST00000644821.1
|
EDN3
|
endothelin 3 |
| chr11_+_124919244 | 0.39 |
ENST00000408930.6
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1 |
| chr7_+_80646347 | 0.39 |
ENST00000413265.5
|
CD36
|
CD36 molecule |
| chr15_-_93089192 | 0.39 |
ENST00000329082.11
|
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chr7_+_27242700 | 0.38 |
ENST00000222761.7
|
EVX1
|
even-skipped homeobox 1 |
| chr6_-_112254555 | 0.38 |
ENST00000230538.12
ENST00000389463.9 ENST00000368638.5 ENST00000431543.6 ENST00000453937.2 |
LAMA4
|
laminin subunit alpha 4 |
| chr5_+_140855882 | 0.38 |
ENST00000562220.2
ENST00000307360.6 ENST00000506939.6 |
PCDHA10
|
protocadherin alpha 10 |
| chr4_+_112860981 | 0.37 |
ENST00000671704.1
|
ANK2
|
ankyrin 2 |
| chr20_+_59300703 | 0.37 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr2_+_69013282 | 0.37 |
ENST00000409829.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr6_-_112254647 | 0.36 |
ENST00000455073.1
ENST00000522006.5 ENST00000519932.5 |
LAMA4
|
laminin subunit alpha 4 |
| chr10_+_69801874 | 0.36 |
ENST00000357811.8
|
COL13A1
|
collagen type XIII alpha 1 chain |
| chr12_-_48570046 | 0.36 |
ENST00000301046.6
ENST00000549817.1 |
LALBA
|
lactalbumin alpha |
| chr6_-_89217339 | 0.36 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1 |
| chr17_+_44846318 | 0.36 |
ENST00000591513.5
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
| chr12_-_23584600 | 0.35 |
ENST00000396007.6
|
SOX5
|
SRY-box transcription factor 5 |
| chr15_+_43692886 | 0.35 |
ENST00000434505.5
ENST00000411750.5 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr7_+_100015572 | 0.33 |
ENST00000535170.5
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr11_-_41459592 | 0.33 |
ENST00000528697.6
ENST00000530763.5 |
LRRC4C
|
leucine rich repeat containing 4C |
| chr16_+_53208438 | 0.33 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr20_+_59300589 | 0.33 |
ENST00000337938.7
ENST00000371025.7 |
EDN3
|
endothelin 3 |
| chr6_+_155216959 | 0.32 |
ENST00000462408.2
|
TIAM2
|
TIAM Rac1 associated GEF 2 |
| chr7_+_27242796 | 0.32 |
ENST00000496902.7
|
EVX1
|
even-skipped homeobox 1 |
| chr14_+_20768393 | 0.32 |
ENST00000326783.4
|
EDDM3B
|
epididymal protein 3B |
| chr17_-_40966945 | 0.32 |
ENST00000355612.7
|
KRT39
|
keratin 39 |
| chr17_-_40100569 | 0.31 |
ENST00000246672.4
|
NR1D1
|
nuclear receptor subfamily 1 group D member 1 |
| chr6_-_112254485 | 0.31 |
ENST00000521398.5
ENST00000424408.6 ENST00000243219.7 |
LAMA4
|
laminin subunit alpha 4 |
| chr5_-_16742221 | 0.30 |
ENST00000505695.5
|
MYO10
|
myosin X |
| chr3_+_137764296 | 0.30 |
ENST00000306087.3
|
SOX14
|
SRY-box transcription factor 14 |
| chr11_-_35360050 | 0.29 |
ENST00000644868.1
ENST00000643454.1 ENST00000646080.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr1_-_230869564 | 0.29 |
ENST00000470540.5
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr3_+_156120572 | 0.28 |
ENST00000389636.9
ENST00000490337.6 |
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr6_-_32178080 | 0.28 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr14_+_74348440 | 0.28 |
ENST00000256362.5
|
VRTN
|
vertebrae development associated |
| chr5_+_55160161 | 0.28 |
ENST00000296734.6
ENST00000515370.1 ENST00000503787.6 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr10_+_52314272 | 0.28 |
ENST00000373970.4
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr22_-_33572227 | 0.27 |
ENST00000674780.1
|
LARGE1
|
LARGE xylosyl- and glucuronyltransferase 1 |
| chr5_-_161546671 | 0.27 |
ENST00000517547.5
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr11_-_76210736 | 0.26 |
ENST00000529461.1
|
WNT11
|
Wnt family member 11 |
| chr17_+_70104848 | 0.26 |
ENST00000392670.5
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr12_+_70366277 | 0.25 |
ENST00000258111.5
|
KCNMB4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr17_+_82860354 | 0.25 |
ENST00000576996.5
|
TBCD
|
tubulin folding cofactor D |
| chr5_+_175861628 | 0.25 |
ENST00000509837.5
|
CPLX2
|
complexin 2 |
| chr6_-_166167832 | 0.24 |
ENST00000366876.7
|
TBXT
|
T-box transcription factor T |
| chr14_-_21034802 | 0.23 |
ENST00000382951.4
|
RNASE13
|
ribonuclease A family member 13 (inactive) |
| chr3_-_57165332 | 0.23 |
ENST00000296318.12
|
IL17RD
|
interleukin 17 receptor D |
| chr10_-_45307838 | 0.23 |
ENST00000536058.1
|
OR13A1
|
olfactory receptor family 13 subfamily A member 1 |
| chr22_-_32464440 | 0.23 |
ENST00000397450.2
ENST00000397452.5 ENST00000300399.8 |
BPIFC
|
BPI fold containing family C |
| chr6_+_37170133 | 0.23 |
ENST00000373509.6
|
PIM1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr5_-_149551168 | 0.22 |
ENST00000515748.2
ENST00000606719.6 |
CSNK1A1
|
casein kinase 1 alpha 1 |
| chr2_+_219444657 | 0.22 |
ENST00000451076.1
|
SPEG
|
striated muscle enriched protein kinase |
| chr13_+_23570370 | 0.22 |
ENST00000403372.6
ENST00000248484.9 |
TNFRSF19
|
TNF receptor superfamily member 19 |
| chr7_+_100015588 | 0.22 |
ENST00000324306.11
ENST00000426572.5 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr5_+_176388731 | 0.22 |
ENST00000274787.3
|
HIGD2A
|
HIG1 hypoxia inducible domain family member 2A |
| chr17_+_70104991 | 0.22 |
ENST00000587698.5
ENST00000587892.1 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr5_+_102865805 | 0.21 |
ENST00000346918.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_-_15797930 | 0.21 |
ENST00000683139.1
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr8_-_71356653 | 0.21 |
ENST00000388742.8
ENST00000388740.4 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr18_+_10454584 | 0.21 |
ENST00000355285.10
|
APCDD1
|
APC down-regulated 1 |
| chr12_-_123149902 | 0.21 |
ENST00000542210.1
|
PITPNM2
|
phosphatidylinositol transfer protein membrane associated 2 |
| chr1_-_44141631 | 0.21 |
ENST00000634670.1
|
KLF18
|
Kruppel like factor 18 |
| chr4_-_108762535 | 0.20 |
ENST00000512320.1
ENST00000510723.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr7_+_114416286 | 0.20 |
ENST00000635534.1
|
FOXP2
|
forkhead box P2 |
| chr10_+_69801892 | 0.20 |
ENST00000398978.8
ENST00000645393.2 ENST00000354547.7 ENST00000674121.1 ENST00000673842.1 ENST00000520267.5 |
COL13A1
|
collagen type XIII alpha 1 chain |
| chrX_-_112679919 | 0.20 |
ENST00000371968.8
|
LHFPL1
|
LHFPL tetraspan subfamily member 1 |
| chr11_-_40294089 | 0.20 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr15_-_89221558 | 0.19 |
ENST00000268125.10
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr11_-_5234475 | 0.19 |
ENST00000292901.7
ENST00000650601.1 ENST00000417377.1 |
HBD
|
hemoglobin subunit delta |
| chr1_+_61952283 | 0.19 |
ENST00000307297.8
|
PATJ
|
PATJ crumbs cell polarity complex component |
| chr4_-_138242325 | 0.18 |
ENST00000280612.9
|
SLC7A11
|
solute carrier family 7 member 11 |
| chr5_+_141417659 | 0.18 |
ENST00000398594.4
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.8 | GO:1903631 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 1.2 | 3.5 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.9 | 4.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.6 | 3.0 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.6 | 1.8 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.5 | 1.5 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.4 | 1.2 | GO:0090381 | regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.4 | 1.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.4 | 1.1 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.3 | 1.3 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.3 | 2.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.3 | 4.8 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 3.0 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 2.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.2 | 3.9 | GO:0070141 | Leydig cell differentiation(GO:0033327) positive regulation of mammary gland epithelial cell proliferation(GO:0033601) response to UV-A(GO:0070141) |
| 0.2 | 0.9 | GO:1905069 | nephrogenic mesenchyme morphogenesis(GO:0072134) allantois development(GO:1905069) |
| 0.2 | 1.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 1.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 3.0 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 0.6 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.2 | 0.6 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.2 | 4.6 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.2 | 0.7 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 1.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 1.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 0.9 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.2 | 3.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 1.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.8 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 1.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.3 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.1 | 0.7 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.7 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.7 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 2.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.4 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.3 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.1 | 6.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.7 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 1.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 2.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.7 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 1.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.2 | GO:0021758 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.1 | 0.8 | GO:2000332 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.9 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 1.0 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 1.5 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 1.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.4 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.9 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 2.5 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.7 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.7 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 1.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 2.9 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 2.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 1.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 1.4 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.3 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 0.9 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.2 | 4.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 2.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 0.6 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 3.0 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 3.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 2.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 3.8 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 1.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 3.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 8.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 1.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.0 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0005816 | spindle pole body(GO:0005816) |
| 0.0 | 1.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 3.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.8 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 1.2 | 8.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.9 | 4.3 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.5 | 2.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.5 | 3.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.4 | 1.3 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.3 | 1.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 1.0 | GO:0030379 | neurotensin receptor activity, non-G-protein coupled(GO:0030379) |
| 0.2 | 0.6 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.2 | 0.7 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.2 | 1.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 1.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.7 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 2.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 2.5 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.7 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 3.0 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 3.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 3.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 1.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 2.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.2 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.1 | 0.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.3 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 2.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 8.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.6 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.9 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.8 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 3.9 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 1.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.0 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 1.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 2.3 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 3.5 | GO:0051015 | actin filament binding(GO:0051015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 3.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 5.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 8.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 3.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 3.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 3.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 3.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 4.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 2.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 4.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |