Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for TEAD4
Z-value: 1.35
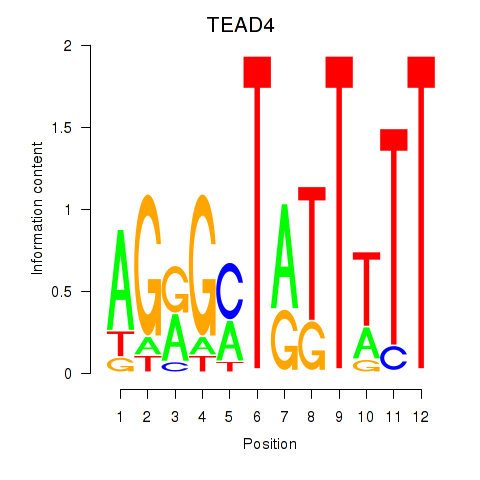
Transcription factors associated with TEAD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD4
|
ENSG00000197905.10 | TEA domain transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD4 | hg38_v1_chr12_+_2959870_2959953 | 0.23 | 2.1e-01 | Click! |
Activity profile of TEAD4 motif
Sorted Z-values of TEAD4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_106335613 | 4.48 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr14_-_106269133 | 4.38 |
ENST00000390609.3
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr14_-_106360320 | 4.23 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr12_+_85874287 | 4.12 |
ENST00000551529.5
ENST00000256010.7 |
NTS
|
neurotensin |
| chr14_-_106593319 | 4.01 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr17_-_31297231 | 3.60 |
ENST00000247271.5
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr14_-_106062670 | 3.24 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr14_-_105987068 | 3.10 |
ENST00000390594.3
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr10_-_73651013 | 2.89 |
ENST00000372873.8
|
SYNPO2L
|
synaptopodin 2 like |
| chr14_-_106165730 | 2.59 |
ENST00000390604.2
|
IGHV3-16
|
immunoglobulin heavy variable 3-16 (non-functional) |
| chr2_-_157327699 | 2.49 |
ENST00000397283.6
|
ERMN
|
ermin |
| chr2_-_89160329 | 2.41 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr14_-_106803221 | 2.35 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr4_-_39032343 | 2.25 |
ENST00000381938.4
|
TMEM156
|
transmembrane protein 156 |
| chr2_-_234497035 | 2.16 |
ENST00000390645.2
ENST00000339728.6 |
ARL4C
|
ADP ribosylation factor like GTPase 4C |
| chr17_+_34255274 | 2.13 |
ENST00000580907.5
ENST00000225831.4 |
CCL2
|
C-C motif chemokine ligand 2 |
| chr16_+_32995048 | 2.09 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr14_-_106088573 | 1.86 |
ENST00000632099.1
|
IGHV3-64D
|
immunoglobulin heavy variable 3-64D |
| chr16_+_32847692 | 1.84 |
ENST00000567458.2
|
IGHV2OR16-5
|
immunoglobulin heavy variable 2/OR16-5 (non-functional) |
| chr6_-_32666648 | 1.84 |
ENST00000399082.7
ENST00000399079.7 ENST00000374943.8 ENST00000434651.6 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr12_+_6444932 | 1.81 |
ENST00000266557.4
|
CD27
|
CD27 molecule |
| chr11_+_60455839 | 1.77 |
ENST00000532491.5
ENST00000532073.5 ENST00000345732.9 ENST00000534668.6 ENST00000528313.1 ENST00000533306.6 ENST00000674194.1 |
MS4A1
|
membrane spanning 4-domains A1 |
| chr16_+_33009175 | 1.62 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr5_+_54455661 | 1.58 |
ENST00000302005.3
|
HSPB3
|
heat shock protein family B (small) member 3 |
| chr1_+_65992389 | 1.51 |
ENST00000423207.6
|
PDE4B
|
phosphodiesterase 4B |
| chr1_+_160796157 | 1.50 |
ENST00000263285.11
ENST00000368039.2 |
LY9
|
lymphocyte antigen 9 |
| chr4_+_41612892 | 1.45 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_119135825 | 1.44 |
ENST00000307128.6
|
MYOZ2
|
myozenin 2 |
| chr13_-_67230377 | 1.43 |
ENST00000544246.5
ENST00000377861.4 |
PCDH9
|
protocadherin 9 |
| chr3_+_130560334 | 1.43 |
ENST00000358511.10
|
COL6A6
|
collagen type VI alpha 6 chain |
| chr3_-_52452828 | 1.38 |
ENST00000496590.1
|
TNNC1
|
troponin C1, slow skeletal and cardiac type |
| chr8_-_27258386 | 1.38 |
ENST00000350889.8
ENST00000519997.5 ENST00000519614.5 ENST00000522908.1 ENST00000265770.11 |
STMN4
|
stathmin 4 |
| chr2_-_178807415 | 1.34 |
ENST00000342992.10
ENST00000460472.6 ENST00000589042.5 ENST00000591111.5 ENST00000360870.10 |
TTN
|
titin |
| chr14_-_106658251 | 1.33 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr20_+_44531817 | 1.32 |
ENST00000372889.5
ENST00000372887.5 |
PKIG
|
cAMP-dependent protein kinase inhibitor gamma |
| chr8_-_27258414 | 1.29 |
ENST00000523048.5
|
STMN4
|
stathmin 4 |
| chr3_+_101827982 | 1.28 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr2_-_136118142 | 1.28 |
ENST00000241393.4
|
CXCR4
|
C-X-C motif chemokine receptor 4 |
| chr10_+_62049211 | 1.27 |
ENST00000309334.5
|
ARID5B
|
AT-rich interaction domain 5B |
| chr1_-_11848345 | 1.25 |
ENST00000376476.1
|
NPPA
|
natriuretic peptide A |
| chr2_+_33476640 | 1.23 |
ENST00000425210.5
ENST00000444784.5 ENST00000423159.5 ENST00000403687.8 |
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr2_+_227813834 | 1.23 |
ENST00000358813.5
ENST00000409189.7 |
CCL20
|
C-C motif chemokine ligand 20 |
| chr1_+_161706949 | 1.22 |
ENST00000350710.3
ENST00000367949.6 ENST00000367959.6 ENST00000540521.5 ENST00000546024.5 ENST00000674251.1 ENST00000674323.1 |
FCRLA
|
Fc receptor like A |
| chr2_+_90021567 | 1.21 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr7_+_54542393 | 1.19 |
ENST00000404951.5
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr2_-_42361149 | 1.19 |
ENST00000468711.5
ENST00000463055.1 |
COX7A2L
|
cytochrome c oxidase subunit 7A2 like |
| chr1_-_101996919 | 1.16 |
ENST00000370103.9
|
OLFM3
|
olfactomedin 3 |
| chrX_+_21374608 | 1.16 |
ENST00000644295.1
ENST00000645074.1 ENST00000645791.1 ENST00000643220.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr12_+_12725897 | 1.15 |
ENST00000326765.10
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr13_-_67230313 | 1.14 |
ENST00000377865.7
|
PCDH9
|
protocadherin 9 |
| chr1_+_160081529 | 1.13 |
ENST00000368088.4
|
KCNJ9
|
potassium inwardly rectifying channel subfamily J member 9 |
| chr19_+_9087061 | 1.13 |
ENST00000641627.1
|
OR1M1
|
olfactory receptor family 1 subfamily M member 1 |
| chr6_-_159726871 | 1.13 |
ENST00000535561.5
|
SOD2
|
superoxide dismutase 2 |
| chr12_+_10307818 | 1.12 |
ENST00000350274.9
ENST00000336164.9 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr6_-_56628018 | 1.11 |
ENST00000522360.5
|
DST
|
dystonin |
| chr7_+_54542362 | 1.10 |
ENST00000402613.4
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr20_-_57359491 | 1.10 |
ENST00000543500.3
|
MTRNR2L3
|
MT-RNR2 like 3 |
| chr6_-_46325641 | 1.10 |
ENST00000330430.10
ENST00000405162.2 |
RCAN2
|
regulator of calcineurin 2 |
| chrX_+_21374357 | 1.07 |
ENST00000643841.1
ENST00000379510.5 ENST00000425654.7 ENST00000644798.1 ENST00000543067.6 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr2_-_60553618 | 1.06 |
ENST00000643716.1
ENST00000359629.10 ENST00000642384.2 ENST00000335712.11 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr3_-_47891269 | 1.04 |
ENST00000335271.9
|
MAP4
|
microtubule associated protein 4 |
| chr2_+_33458043 | 1.04 |
ENST00000437184.5
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr11_+_94706412 | 1.04 |
ENST00000299004.13
|
AMOTL1
|
angiomotin like 1 |
| chr12_-_91146195 | 1.03 |
ENST00000548218.1
|
DCN
|
decorin |
| chr20_+_58841612 | 1.02 |
ENST00000462499.6
ENST00000482112.6 ENST00000663479.2 ENST00000493744.5 |
GNAS
|
GNAS complex locus |
| chr4_+_41612702 | 1.01 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr16_-_46763237 | 1.00 |
ENST00000536476.5
|
MYLK3
|
myosin light chain kinase 3 |
| chr7_-_111788958 | 1.00 |
ENST00000417165.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr8_+_66493514 | 1.00 |
ENST00000521495.5
|
VXN
|
vexin |
| chr2_+_200440649 | 0.99 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr7_-_36985060 | 0.97 |
ENST00000396040.6
|
ELMO1
|
engulfment and cell motility 1 |
| chr1_+_163068775 | 0.97 |
ENST00000421743.6
|
RGS4
|
regulator of G protein signaling 4 |
| chr7_+_87934242 | 0.95 |
ENST00000413139.2
ENST00000412441.5 ENST00000683525.1 ENST00000398201.8 ENST00000265727.11 ENST00000398209.7 ENST00000684002.1 |
ADAM22
|
ADAM metallopeptidase domain 22 |
| chrX_+_21374476 | 0.94 |
ENST00000644585.1
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chrX_+_21374434 | 0.91 |
ENST00000279451.9
ENST00000645245.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr12_-_9733292 | 0.90 |
ENST00000621400.4
ENST00000327839.3 |
CLECL1
|
C-type lectin like 1 |
| chr2_+_33476616 | 0.90 |
ENST00000442390.5
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr7_+_54542300 | 0.90 |
ENST00000302287.7
ENST00000407838.7 |
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr7_-_83648547 | 0.90 |
ENST00000642232.1
|
SEMA3E
|
semaphorin 3E |
| chr3_+_63443306 | 0.88 |
ENST00000472899.5
ENST00000479198.5 ENST00000460711.5 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr2_-_60553558 | 0.88 |
ENST00000642439.1
ENST00000356842.9 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr1_+_32251239 | 0.88 |
ENST00000373564.7
ENST00000482949.5 ENST00000336890.10 ENST00000495610.6 |
LCK
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr12_-_48999363 | 0.87 |
ENST00000421952.3
|
DDN
|
dendrin |
| chr19_+_35139724 | 0.87 |
ENST00000588715.5
ENST00000588607.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr5_+_58583068 | 0.87 |
ENST00000282878.6
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr7_-_22194709 | 0.86 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr15_-_19988117 | 0.86 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr10_-_59753444 | 0.85 |
ENST00000594536.5
ENST00000414264.6 |
MRLN
|
myoregulin |
| chr18_+_54591159 | 0.84 |
ENST00000321600.1
ENST00000648945.2 |
DYNAP
|
dynactin associated protein |
| chr6_-_46015812 | 0.84 |
ENST00000544153.3
ENST00000339561.12 |
CLIC5
|
chloride intracellular channel 5 |
| chr12_-_9760893 | 0.83 |
ENST00000228434.7
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr2_-_197676012 | 0.83 |
ENST00000429081.1
|
RFTN2
|
raftlin family member 2 |
| chr7_+_148133684 | 0.82 |
ENST00000628930.2
|
CNTNAP2
|
contactin associated protein 2 |
| chr3_+_124094663 | 0.82 |
ENST00000460856.5
ENST00000240874.7 |
KALRN
|
kalirin RhoGEF kinase |
| chr3_+_63443076 | 0.82 |
ENST00000295894.9
|
SYNPR
|
synaptoporin |
| chr15_+_33968484 | 0.81 |
ENST00000383263.7
|
CHRM5
|
cholinergic receptor muscarinic 5 |
| chr4_+_185395979 | 0.81 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr19_+_35138993 | 0.80 |
ENST00000612146.4
ENST00000589209.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr1_+_43270007 | 0.79 |
ENST00000432792.6
|
TMEM125
|
transmembrane protein 125 |
| chr6_+_31670167 | 0.78 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 family member G5B |
| chr7_+_142313144 | 0.78 |
ENST00000390357.3
|
TRBV4-1
|
T cell receptor beta variable 4-1 |
| chrX_+_21374288 | 0.77 |
ENST00000642359.1
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr6_-_56642788 | 0.77 |
ENST00000439203.5
ENST00000518935.5 ENST00000370765.11 ENST00000244364.10 |
DST
|
dystonin |
| chr8_+_66493556 | 0.77 |
ENST00000305454.8
ENST00000522977.5 ENST00000480005.1 |
VXN
|
vexin |
| chr19_-_51417619 | 0.75 |
ENST00000441969.7
ENST00000339313.10 ENST00000525998.5 ENST00000436984.6 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr5_-_124745315 | 0.75 |
ENST00000306315.9
|
ZNF608
|
zinc finger protein 608 |
| chr5_+_173889337 | 0.75 |
ENST00000520867.5
ENST00000334035.9 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr6_-_134317629 | 0.74 |
ENST00000524929.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr11_+_120385286 | 0.74 |
ENST00000532993.5
|
ARHGEF12
|
Rho guanine nucleotide exchange factor 12 |
| chr10_+_49299159 | 0.72 |
ENST00000374144.8
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr19_+_35139440 | 0.71 |
ENST00000455515.6
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr7_+_151114597 | 0.70 |
ENST00000335367.7
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr1_+_43269974 | 0.70 |
ENST00000439858.6
|
TMEM125
|
transmembrane protein 125 |
| chr5_-_41794211 | 0.70 |
ENST00000512084.5
|
OXCT1
|
3-oxoacid CoA-transferase 1 |
| chr14_-_75063990 | 0.70 |
ENST00000555135.1
ENST00000357971.7 ENST00000553302.1 ENST00000238618.8 ENST00000555694.5 |
ACYP1
|
acylphosphatase 1 |
| chr3_+_124094696 | 0.69 |
ENST00000360013.7
ENST00000684186.1 ENST00000684276.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr18_+_58341038 | 0.69 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr7_-_81770122 | 0.69 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor |
| chr2_-_150539011 | 0.68 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr4_+_118034480 | 0.68 |
ENST00000296499.6
|
NDST3
|
N-deacetylase and N-sulfotransferase 3 |
| chr8_-_23854796 | 0.68 |
ENST00000290271.7
|
STC1
|
stanniocalcin 1 |
| chr10_-_59753388 | 0.68 |
ENST00000430431.5
|
MRLN
|
myoregulin |
| chr17_+_4951758 | 0.67 |
ENST00000518175.1
|
ENO3
|
enolase 3 |
| chr15_+_81182579 | 0.67 |
ENST00000302987.9
|
IL16
|
interleukin 16 |
| chr6_-_134317905 | 0.67 |
ENST00000461976.2
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr11_-_78079773 | 0.66 |
ENST00000612612.5
ENST00000614236.2 |
NDUFC2-KCTD14
|
NDUFC2-KCTD14 readthrough |
| chr12_-_122526929 | 0.64 |
ENST00000331738.12
ENST00000528279.1 ENST00000344591.8 ENST00000526560.6 |
RSRC2
|
arginine and serine rich coiled-coil 2 |
| chr4_+_146214515 | 0.63 |
ENST00000636502.1
|
REELD1
|
reeler domain containing 1 |
| chr21_+_33432173 | 0.63 |
ENST00000421802.1
|
IFNGR2
|
interferon gamma receptor 2 |
| chr15_+_80441229 | 0.62 |
ENST00000533983.5
ENST00000527771.5 ENST00000525103.1 |
ARNT2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr3_-_100847647 | 0.62 |
ENST00000528490.5
|
ABI3BP
|
ABI family member 3 binding protein |
| chr10_-_60389833 | 0.61 |
ENST00000280772.7
|
ANK3
|
ankyrin 3 |
| chr5_-_41794547 | 0.61 |
ENST00000510634.5
|
OXCT1
|
3-oxoacid CoA-transferase 1 |
| chrX_-_32412220 | 0.60 |
ENST00000619831.5
|
DMD
|
dystrophin |
| chr2_+_209771972 | 0.60 |
ENST00000439458.5
ENST00000272845.10 |
UNC80
|
unc-80 homolog, NALCN channel complex subunit |
| chr8_+_49911604 | 0.60 |
ENST00000642164.1
ENST00000644093.1 ENST00000643999.1 ENST00000647073.1 ENST00000646880.1 |
SNTG1
|
syntrophin gamma 1 |
| chr11_-_102724781 | 0.60 |
ENST00000438475.2
|
MMP8
|
matrix metallopeptidase 8 |
| chr4_+_36281591 | 0.59 |
ENST00000639862.2
ENST00000357504.7 |
DTHD1
|
death domain containing 1 |
| chr3_-_194633689 | 0.59 |
ENST00000330115.3
|
TMEM44
|
transmembrane protein 44 |
| chr7_-_82443715 | 0.58 |
ENST00000356253.9
ENST00000423588.1 |
CACNA2D1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr7_-_81769971 | 0.58 |
ENST00000354224.10
ENST00000643024.1 |
HGF
|
hepatocyte growth factor |
| chr5_+_174045673 | 0.58 |
ENST00000303177.8
ENST00000519867.5 |
NSG2
|
neuronal vesicle trafficking associated 2 |
| chr9_+_74497308 | 0.58 |
ENST00000376896.8
|
RORB
|
RAR related orphan receptor B |
| chr3_-_18424533 | 0.57 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr12_+_32107296 | 0.57 |
ENST00000551086.1
|
BICD1
|
BICD cargo adaptor 1 |
| chr10_+_122163672 | 0.57 |
ENST00000369004.7
ENST00000260733.7 |
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr12_-_39619782 | 0.57 |
ENST00000308666.4
|
ABCD2
|
ATP binding cassette subfamily D member 2 |
| chr20_+_34977625 | 0.56 |
ENST00000618182.6
|
MYH7B
|
myosin heavy chain 7B |
| chr11_+_10455292 | 0.56 |
ENST00000396553.6
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr12_-_9733083 | 0.56 |
ENST00000542530.5
|
CLECL1
|
C-type lectin like 1 |
| chr8_+_49911396 | 0.55 |
ENST00000642720.2
|
SNTG1
|
syntrophin gamma 1 |
| chr7_-_47948448 | 0.55 |
ENST00000289672.7
|
PKD1L1
|
polycystin 1 like 1, transient receptor potential channel interacting |
| chr12_-_124917340 | 0.55 |
ENST00000542416.1
|
UBC
|
ubiquitin C |
| chr3_-_69200711 | 0.55 |
ENST00000478263.5
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr15_-_38560050 | 0.55 |
ENST00000558432.5
|
RASGRP1
|
RAS guanyl releasing protein 1 |
| chr1_-_27626106 | 0.54 |
ENST00000457296.5
|
FGR
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr6_-_152637386 | 0.54 |
ENST00000610489.2
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr17_-_15272287 | 0.53 |
ENST00000674868.1
|
PMP22
|
peripheral myelin protein 22 |
| chr17_-_36196748 | 0.53 |
ENST00000619989.1
|
CCL3L1
|
C-C motif chemokine ligand 3 like 1 |
| chr18_+_63587336 | 0.53 |
ENST00000344731.10
|
SERPINB13
|
serpin family B member 13 |
| chr3_-_197260369 | 0.53 |
ENST00000658155.1
ENST00000453607.5 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr19_+_9185594 | 0.53 |
ENST00000344248.4
|
OR7D2
|
olfactory receptor family 7 subfamily D member 2 |
| chr18_+_63587297 | 0.52 |
ENST00000269489.9
|
SERPINB13
|
serpin family B member 13 |
| chr1_+_161707205 | 0.52 |
ENST00000367957.7
|
FCRLA
|
Fc receptor like A |
| chr9_-_101487120 | 0.52 |
ENST00000374848.8
|
PGAP4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr6_+_31670379 | 0.52 |
ENST00000409525.1
|
LY6G5B
|
lymphocyte antigen 6 family member G5B |
| chr8_-_69833338 | 0.52 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr4_+_94489030 | 0.50 |
ENST00000510099.5
|
PDLIM5
|
PDZ and LIM domain 5 |
| chr21_-_38498415 | 0.50 |
ENST00000398905.5
ENST00000398907.5 ENST00000453032.6 ENST00000288319.12 |
ERG
|
ETS transcription factor ERG |
| chr2_-_187554473 | 0.50 |
ENST00000453013.5
ENST00000417013.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr11_+_59511539 | 0.50 |
ENST00000641962.1
|
OR4D9
|
olfactory receptor family 4 subfamily D member 9 |
| chr8_-_42502496 | 0.50 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 member 2 |
| chr3_-_54928044 | 0.49 |
ENST00000273286.6
|
LRTM1
|
leucine rich repeats and transmembrane domains 1 |
| chr19_-_3786408 | 0.49 |
ENST00000395040.6
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr4_+_70430487 | 0.49 |
ENST00000413702.5
|
MUC7
|
mucin 7, secreted |
| chr2_+_190927649 | 0.48 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr9_-_73865 | 0.48 |
ENST00000642633.1
|
WASHC1
|
WASH complex subunit 1 |
| chr1_-_211492111 | 0.48 |
ENST00000367002.5
ENST00000680073.1 |
RD3
|
RD3 regulator of GUCY2D |
| chr6_-_27867581 | 0.48 |
ENST00000331442.5
|
H1-5
|
H1.5 linker histone, cluster member |
| chr22_-_50085378 | 0.47 |
ENST00000442311.1
|
MLC1
|
modulator of VRAC current 1 |
| chr2_-_44323302 | 0.47 |
ENST00000420756.1
ENST00000444696.5 |
PREPL
|
prolyl endopeptidase like |
| chr13_-_44437214 | 0.47 |
ENST00000622051.1
|
TSC22D1
|
TSC22 domain family member 1 |
| chr4_+_153222402 | 0.47 |
ENST00000676335.1
ENST00000675146.1 |
TRIM2
|
tripartite motif containing 2 |
| chr17_+_46713149 | 0.47 |
ENST00000576346.5
|
NSF
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr10_-_113664033 | 0.46 |
ENST00000359988.4
ENST00000369360.7 ENST00000360478.7 ENST00000369358.8 |
NRAP
|
nebulin related anchoring protein |
| chr1_+_15684284 | 0.46 |
ENST00000375799.8
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology and RUN domain containing M2 |
| chr1_+_161707222 | 0.46 |
ENST00000236938.12
|
FCRLA
|
Fc receptor like A |
| chr7_-_13989658 | 0.46 |
ENST00000430479.6
ENST00000433547.1 ENST00000405192.6 |
ETV1
|
ETS variant transcription factor 1 |
| chr6_-_152168349 | 0.46 |
ENST00000539504.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr12_+_10307950 | 0.46 |
ENST00000543420.5
ENST00000543777.5 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr19_-_3786363 | 0.46 |
ENST00000310132.11
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr19_-_3786254 | 0.46 |
ENST00000585778.5
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr4_-_20984011 | 0.46 |
ENST00000382149.9
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr12_+_108129276 | 0.46 |
ENST00000547525.6
|
WSCD2
|
WSC domain containing 2 |
| chr17_-_18682262 | 0.45 |
ENST00000454745.2
ENST00000395675.7 |
FOXO3B
|
forkhead box O3B |
| chr8_+_24294044 | 0.45 |
ENST00000265769.9
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr2_-_189225175 | 0.45 |
ENST00000649966.1
|
COL5A2
|
collagen type V alpha 2 chain |
| chr6_-_154247630 | 0.45 |
ENST00000519344.5
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr5_-_139482714 | 0.45 |
ENST00000652543.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1 |
| chr8_-_133499320 | 0.44 |
ENST00000523855.1
ENST00000523854.5 |
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.6 | 3.2 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.4 | 2.2 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.4 | 1.8 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.4 | 1.1 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.3 | 1.0 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.3 | 1.4 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 2.4 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.3 | 1.2 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.3 | 1.5 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.3 | 2.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.2 | 1.1 | GO:0003070 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 0.9 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.2 | 1.2 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.2 | 17.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 1.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 1.6 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 | 17.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 1.3 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 1.5 | GO:0046959 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 1.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.6 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.1 | 0.8 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.4 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 1.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.5 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.3 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.6 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 1.8 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) |
| 0.1 | 1.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.6 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.5 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 0.6 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.4 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.8 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 0.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 1.5 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.1 | 1.4 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.7 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.4 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.1 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 1.0 | GO:2000828 | post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.1 | 0.6 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.5 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.1 | 1.0 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.5 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.8 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0032258 | CVT pathway(GO:0032258) G-protein coupled receptor catabolic process(GO:1990172) |
| 0.0 | 0.4 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.3 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.2 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.4 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 2.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.4 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.9 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.8 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.5 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.8 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.4 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 2.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 3.6 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.8 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 2.9 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.7 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.9 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.8 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 1.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.5 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.9 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.1 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.8 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.6 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.0 | 1.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 1.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 2.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.8 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 1.9 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 2.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.5 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 17.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 1.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 0.9 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.2 | 1.9 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 0.8 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.2 | 2.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 0.5 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.1 | 4.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 2.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.4 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 1.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 2.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.4 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 1.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.3 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 1.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 1.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.5 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 4.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 10.4 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 5.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 4.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 4.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 2.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 1.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.3 | 1.6 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.3 | 1.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 17.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.6 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.2 | 1.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 1.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 5.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.9 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.6 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 3.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 26.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.4 | GO:0004948 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.1 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.3 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.7 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.1 | 0.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 2.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.3 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 1.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 4.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.0 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.0 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 1.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 1.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 2.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0090079 | translation activator activity(GO:0008494) translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.7 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 7.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 3.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 3.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 3.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.6 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 3.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 6.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 3.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 3.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.6 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 1.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |