Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
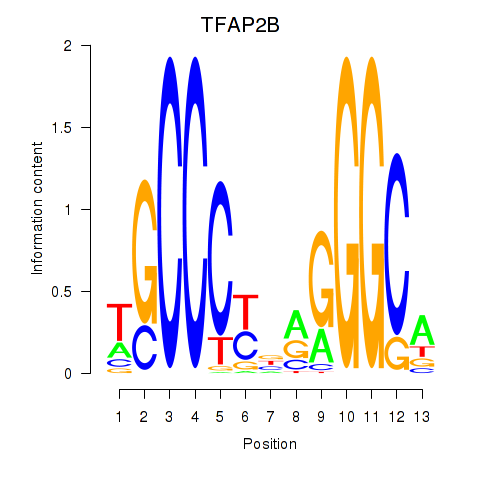
Results for TFAP2B
Z-value: 1.00
Transcription factors associated with TFAP2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2B
|
ENSG00000008196.13 | transcription factor AP-2 beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFAP2B | hg38_v1_chr6_+_50818701_50818723 | 0.02 | 9.2e-01 | Click! |
Activity profile of TFAP2B motif
Sorted Z-values of TFAP2B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_63700100 | 5.89 |
ENST00000578993.5
ENST00000259006.8 ENST00000583211.5 |
LIMD2
|
LIM domain containing 2 |
| chr4_+_1721470 | 4.38 |
ENST00000612220.5
ENST00000313288.9 |
TACC3
|
transforming acidic coiled-coil containing protein 3 |
| chr4_-_83109843 | 3.35 |
ENST00000411416.6
|
PLAC8
|
placenta associated 8 |
| chr5_-_157575767 | 3.22 |
ENST00000257527.9
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr16_-_88706353 | 3.18 |
ENST00000567844.1
ENST00000312838.9 |
RNF166
|
ring finger protein 166 |
| chr1_+_209756032 | 3.10 |
ENST00000400959.7
ENST00000367025.8 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr1_+_32251239 | 2.83 |
ENST00000373564.7
ENST00000482949.5 ENST00000336890.10 ENST00000495610.6 |
LCK
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr19_+_7645895 | 2.81 |
ENST00000602355.1
|
STXBP2
|
syntaxin binding protein 2 |
| chr1_+_206507520 | 2.65 |
ENST00000579436.7
|
RASSF5
|
Ras association domain family member 5 |
| chr16_-_88706262 | 2.63 |
ENST00000562544.1
|
RNF166
|
ring finger protein 166 |
| chr1_+_206507546 | 2.62 |
ENST00000580449.5
ENST00000581503.6 |
RASSF5
|
Ras association domain family member 5 |
| chr1_+_209756149 | 2.61 |
ENST00000367026.7
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr19_-_19628197 | 2.56 |
ENST00000586703.1
ENST00000591042.1 ENST00000407877.8 |
LPAR2
|
lysophosphatidic acid receptor 2 |
| chrX_+_132023294 | 2.53 |
ENST00000481105.5
ENST00000354719.10 ENST00000394334.7 ENST00000394335.6 |
STK26
|
serine/threonine kinase 26 |
| chr5_-_157575741 | 2.53 |
ENST00000517905.1
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr7_+_99374722 | 2.47 |
ENST00000418347.6
ENST00000429246.6 ENST00000417330.6 ENST00000431816.6 ENST00000458033.6 ENST00000451682.5 ENST00000427217.6 ENST00000646101.2 |
ARPC1B
|
actin related protein 2/3 complex subunit 1B |
| chr8_+_38901327 | 2.46 |
ENST00000519640.5
ENST00000617275.5 |
PLEKHA2
|
pleckstrin homology domain containing A2 |
| chr7_+_99374675 | 2.43 |
ENST00000645391.1
ENST00000455009.6 |
ARPC1B
|
actin related protein 2/3 complex subunit 1B |
| chr11_+_64206663 | 2.33 |
ENST00000544997.5
ENST00000345728.10 ENST00000279227.9 |
FERMT3
|
fermitin family member 3 |
| chr16_+_23835946 | 2.29 |
ENST00000321728.12
ENST00000643927.1 |
PRKCB
|
protein kinase C beta |
| chr12_-_89352395 | 2.28 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr8_+_38901218 | 2.18 |
ENST00000521746.5
ENST00000616927.4 |
PLEKHA2
|
pleckstrin homology domain containing A2 |
| chr12_-_89352487 | 2.16 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr20_-_48827992 | 2.15 |
ENST00000371941.4
|
PREX1
|
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 1 |
| chr1_+_172659095 | 2.13 |
ENST00000367721.3
ENST00000340030.4 |
FASLG
|
Fas ligand |
| chr19_-_10339610 | 2.12 |
ENST00000589261.5
ENST00000160262.10 ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr19_+_7637099 | 1.99 |
ENST00000595950.5
ENST00000221283.10 ENST00000441779.6 ENST00000414284.6 |
STXBP2
|
syntaxin binding protein 2 |
| chr6_-_30687200 | 1.99 |
ENST00000399199.7
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr17_+_44352109 | 1.98 |
ENST00000586242.1
|
GRN
|
granulin precursor |
| chr2_+_85695368 | 1.95 |
ENST00000526018.1
|
GNLY
|
granulysin |
| chr14_+_92513766 | 1.95 |
ENST00000216487.12
ENST00000620541.4 ENST00000557762.1 |
RIN3
|
Ras and Rab interactor 3 |
| chr1_-_24964984 | 1.94 |
ENST00000338888.3
ENST00000399916.5 |
RUNX3
|
RUNX family transcription factor 3 |
| chr17_-_63699730 | 1.92 |
ENST00000578061.5
|
LIMD2
|
LIM domain containing 2 |
| chrX_+_78945332 | 1.91 |
ENST00000544091.1
ENST00000171757.3 |
P2RY10
|
P2Y receptor family member 10 |
| chr19_-_11578937 | 1.91 |
ENST00000592659.1
ENST00000592828.6 ENST00000218758.9 ENST00000412435.6 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr6_-_30686624 | 1.90 |
ENST00000274853.8
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr12_+_6951345 | 1.85 |
ENST00000318974.14
ENST00000541698.5 ENST00000542462.1 |
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chrX_+_132023580 | 1.79 |
ENST00000496850.1
|
STK26
|
serine/threonine kinase 26 |
| chr17_-_45425620 | 1.77 |
ENST00000376922.6
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr19_+_35154715 | 1.76 |
ENST00000392218.6
ENST00000543307.5 ENST00000392219.7 ENST00000541435.6 ENST00000590686.5 ENST00000342879.7 ENST00000588699.5 |
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr11_-_417385 | 1.76 |
ENST00000332725.7
|
SIGIRR
|
single Ig and TIR domain containing |
| chr1_-_97920986 | 1.74 |
ENST00000370192.8
|
DPYD
|
dihydropyrimidine dehydrogenase |
| chr17_-_5234801 | 1.68 |
ENST00000571800.5
ENST00000574081.6 ENST00000399600.8 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr12_+_6951271 | 1.67 |
ENST00000456013.5
|
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chr1_+_155208690 | 1.67 |
ENST00000368376.8
|
MTX1
|
metaxin 1 |
| chr10_+_87863595 | 1.67 |
ENST00000371953.8
|
PTEN
|
phosphatase and tensin homolog |
| chr17_+_74737211 | 1.65 |
ENST00000392612.7
ENST00000392610.5 |
RAB37
|
RAB37, member RAS oncogene family |
| chr7_-_150801325 | 1.65 |
ENST00000447204.6
|
TMEM176B
|
transmembrane protein 176B |
| chr7_+_150801522 | 1.62 |
ENST00000461345.5
|
TMEM176A
|
transmembrane protein 176A |
| chr17_+_74737238 | 1.60 |
ENST00000392613.10
ENST00000613645.1 |
RAB37
|
RAB37, member RAS oncogene family |
| chr16_+_81444799 | 1.60 |
ENST00000537098.8
|
CMIP
|
c-Maf inducing protein |
| chr16_+_28984795 | 1.59 |
ENST00000395461.7
|
LAT
|
linker for activation of T cells |
| chr14_+_21736136 | 1.57 |
ENST00000390426.2
|
TRAV4
|
T cell receptor alpha variable 4 |
| chr7_-_87220567 | 1.50 |
ENST00000433078.5
|
TMEM243
|
transmembrane protein 243 |
| chr7_+_150801695 | 1.50 |
ENST00000475536.5
ENST00000468689.2 |
TMEM176A
|
transmembrane protein 176A |
| chr19_+_44748673 | 1.46 |
ENST00000164227.10
|
BCL3
|
BCL3 transcription coactivator |
| chr22_-_30266839 | 1.44 |
ENST00000403463.1
ENST00000215781.3 |
OSM
|
oncostatin M |
| chr1_+_209756101 | 1.43 |
ENST00000479796.5
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr19_+_35154914 | 1.43 |
ENST00000423817.7
|
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr12_+_6951250 | 1.40 |
ENST00000538715.5
|
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chrX_-_19887585 | 1.40 |
ENST00000397821.8
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr19_+_1067272 | 1.39 |
ENST00000590214.5
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr3_+_158732183 | 1.39 |
ENST00000486568.5
ENST00000495318.5 |
MFSD1
ENSG00000240207.7
|
major facilitator superfamily domain containing 1 novel transcript, antisense to RARRES1 |
| chr5_+_56815534 | 1.39 |
ENST00000399503.4
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr19_+_1067493 | 1.38 |
ENST00000586866.5
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr22_+_19723525 | 1.38 |
ENST00000366425.4
|
GP1BB
|
glycoprotein Ib platelet subunit beta |
| chr19_+_1067144 | 1.37 |
ENST00000313093.7
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr17_-_5584448 | 1.35 |
ENST00000269280.8
ENST00000571451.6 ENST00000572272.6 ENST00000613500.4 ENST00000619223.4 ENST00000617618.4 ENST00000345221.7 ENST00000262467.10 |
NLRP1
|
NLR family pyrin domain containing 1 |
| chr12_+_68610858 | 1.31 |
ENST00000393436.9
ENST00000425247.6 ENST00000250559.14 ENST00000489473.6 ENST00000422358.6 ENST00000541167.5 ENST00000538283.5 ENST00000341355.9 ENST00000537460.5 ENST00000450214.6 ENST00000545270.5 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr5_+_177303768 | 1.31 |
ENST00000303204.9
ENST00000503216.5 |
PRELID1
|
PRELI domain containing 1 |
| chr1_+_155209213 | 1.31 |
ENST00000609421.1
|
MTX1
|
metaxin 1 |
| chr8_-_100721851 | 1.30 |
ENST00000522658.6
|
PABPC1
|
poly(A) binding protein cytoplasmic 1 |
| chr1_-_212699817 | 1.30 |
ENST00000243440.2
|
BATF3
|
basic leucine zipper ATF-like transcription factor 3 |
| chr2_+_55232359 | 1.30 |
ENST00000449323.5
|
RPS27A
|
ribosomal protein S27a |
| chr8_-_100721942 | 1.29 |
ENST00000522387.5
|
PABPC1
|
poly(A) binding protein cytoplasmic 1 |
| chr1_+_155208727 | 1.29 |
ENST00000316721.8
|
MTX1
|
metaxin 1 |
| chr1_+_100352451 | 1.27 |
ENST00000361544.11
ENST00000370124.8 ENST00000336454.5 |
CDC14A
|
cell division cycle 14A |
| chr7_+_98211431 | 1.24 |
ENST00000609256.2
|
BHLHA15
|
basic helix-loop-helix family member a15 |
| chr1_+_28887072 | 1.22 |
ENST00000647103.1
ENST00000642937.2 ENST00000646189.1 ENST00000644342.1 ENST00000373800.7 ENST00000649717.1 |
EPB41
|
erythrocyte membrane protein band 4.1 |
| chr2_-_207165923 | 1.22 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr3_+_53161120 | 1.21 |
ENST00000394729.6
ENST00000330452.8 ENST00000652449.1 |
PRKCD
|
protein kinase C delta |
| chr11_-_6655788 | 1.21 |
ENST00000299441.5
|
DCHS1
|
dachsous cadherin-related 1 |
| chr8_-_100722036 | 1.19 |
ENST00000518196.5
ENST00000519004.5 ENST00000519363.1 ENST00000318607.10 ENST00000520142.2 |
PABPC1
|
poly(A) binding protein cytoplasmic 1 |
| chr1_-_161069666 | 1.19 |
ENST00000368016.7
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr1_+_28887166 | 1.19 |
ENST00000347529.7
|
EPB41
|
erythrocyte membrane protein band 4.1 |
| chr10_+_102419189 | 1.18 |
ENST00000432590.5
|
FBXL15
|
F-box and leucine rich repeat protein 15 |
| chr19_+_37907200 | 1.17 |
ENST00000222345.11
|
SIPA1L3
|
signal induced proliferation associated 1 like 3 |
| chr11_-_47378527 | 1.17 |
ENST00000378538.8
|
SPI1
|
Spi-1 proto-oncogene |
| chr10_+_48684859 | 1.16 |
ENST00000360890.6
ENST00000325239.11 |
WDFY4
|
WDFY family member 4 |
| chr11_-_47378391 | 1.15 |
ENST00000227163.8
|
SPI1
|
Spi-1 proto-oncogene |
| chr19_-_42220109 | 1.15 |
ENST00000595337.5
|
DEDD2
|
death effector domain containing 2 |
| chr1_+_153774210 | 1.14 |
ENST00000271857.6
|
SLC27A3
|
solute carrier family 27 member 3 |
| chr16_-_67936808 | 1.13 |
ENST00000358514.9
|
PSMB10
|
proteasome 20S subunit beta 10 |
| chrX_+_49171889 | 1.12 |
ENST00000376327.6
|
PLP2
|
proteolipid protein 2 |
| chr1_+_27959943 | 1.12 |
ENST00000675575.1
ENST00000373884.6 |
XKR8
|
XK related 8 |
| chrX_+_49171918 | 1.12 |
ENST00000376322.7
|
PLP2
|
proteolipid protein 2 |
| chr19_-_55325316 | 1.11 |
ENST00000591570.5
ENST00000326652.9 |
TMEM150B
|
transmembrane protein 150B |
| chrX_-_19887459 | 1.10 |
ENST00000379697.7
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr11_-_47378494 | 1.10 |
ENST00000533030.1
|
SPI1
|
Spi-1 proto-oncogene |
| chr19_-_1863497 | 1.09 |
ENST00000617223.1
ENST00000250916.6 |
KLF16
|
Kruppel like factor 16 |
| chr12_-_14961610 | 1.09 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr19_-_19628512 | 1.09 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr12_-_6606320 | 1.08 |
ENST00000642594.1
ENST00000644289.1 ENST00000645095.1 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr6_+_33075952 | 1.07 |
ENST00000418931.7
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr12_+_68610956 | 1.07 |
ENST00000545720.6
|
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr12_-_14961559 | 1.06 |
ENST00000228945.9
ENST00000541546.5 |
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr8_+_141128581 | 1.06 |
ENST00000519811.6
|
DENND3
|
DENN domain containing 3 |
| chr11_-_47378479 | 1.05 |
ENST00000533968.1
|
SPI1
|
Spi-1 proto-oncogene |
| chr1_-_161069962 | 1.05 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr15_+_41332862 | 1.05 |
ENST00000450592.6
ENST00000414849.6 ENST00000559596.6 ENST00000560747.5 ENST00000560177.5 |
NUSAP1
|
nucleolar and spindle associated protein 1 |
| chr9_+_87498491 | 1.05 |
ENST00000622514.4
|
DAPK1
|
death associated protein kinase 1 |
| chr1_+_178725227 | 1.04 |
ENST00000367635.8
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr8_+_22578249 | 1.04 |
ENST00000456545.5
|
PDLIM2
|
PDZ and LIM domain 2 |
| chr11_+_810227 | 1.03 |
ENST00000530398.1
|
RPLP2
|
ribosomal protein lateral stalk subunit P2 |
| chr4_-_13544506 | 1.03 |
ENST00000382438.6
|
NKX3-2
|
NK3 homeobox 2 |
| chr1_+_178725277 | 1.01 |
ENST00000324778.5
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr16_-_11256192 | 0.98 |
ENST00000644787.1
ENST00000332029.4 |
SOCS1
|
suppressor of cytokine signaling 1 |
| chr9_+_126860625 | 0.98 |
ENST00000319119.4
|
ZBTB34
|
zinc finger and BTB domain containing 34 |
| chr7_+_7968787 | 0.97 |
ENST00000223145.10
|
GLCCI1
|
glucocorticoid induced 1 |
| chr19_+_55386338 | 0.97 |
ENST00000558131.1
ENST00000558752.1 |
RPL28
|
ribosomal protein L28 |
| chrX_-_154805386 | 0.97 |
ENST00000393531.5
ENST00000369534.8 ENST00000453245.5 ENST00000428488.1 ENST00000369531.1 |
MPP1
|
membrane palmitoylated protein 1 |
| chr2_+_176151543 | 0.96 |
ENST00000306324.4
|
HOXD4
|
homeobox D4 |
| chrX_+_9465011 | 0.96 |
ENST00000645353.2
|
TBL1X
|
transducin beta like 1 X-linked |
| chr1_+_178725147 | 0.95 |
ENST00000367634.6
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr11_+_809961 | 0.95 |
ENST00000321153.9
ENST00000530797.5 |
RPLP2
|
ribosomal protein lateral stalk subunit P2 |
| chr4_-_77819356 | 0.94 |
ENST00000649644.1
ENST00000504123.6 ENST00000515441.2 |
CNOT6L
|
CCR4-NOT transcription complex subunit 6 like |
| chr12_+_68611025 | 0.93 |
ENST00000541216.1
|
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr1_+_40040720 | 0.92 |
ENST00000414893.5
ENST00000372792.7 ENST00000372805.8 ENST00000414281.5 ENST00000420216.5 ENST00000372798.5 ENST00000340450.7 ENST00000435719.5 ENST00000427843.5 ENST00000417287.5 ENST00000424977.1 |
CAP1
|
cyclase associated actin cytoskeleton regulatory protein 1 |
| chr9_-_71768386 | 0.91 |
ENST00000377066.9
ENST00000377044.9 |
CEMIP2
|
cell migration inducing hyaluronidase 2 |
| chr14_+_71586261 | 0.89 |
ENST00000358550.6
|
SIPA1L1
|
signal induced proliferation associated 1 like 1 |
| chr8_-_8893548 | 0.89 |
ENST00000276282.7
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr8_-_100721583 | 0.88 |
ENST00000610907.2
ENST00000677140.1 ENST00000677787.1 ENST00000679197.1 ENST00000519100.6 |
PABPC1
|
poly(A) binding protein cytoplasmic 1 |
| chr1_-_161069857 | 0.88 |
ENST00000368013.8
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr1_-_97921042 | 0.88 |
ENST00000306031.5
|
DPYD
|
dihydropyrimidine dehydrogenase |
| chr15_-_56243829 | 0.88 |
ENST00000559447.8
ENST00000673997.1 |
RFX7
|
regulatory factor X7 |
| chr14_+_75428011 | 0.88 |
ENST00000651602.1
ENST00000559060.5 |
JDP2
|
Jun dimerization protein 2 |
| chr7_-_128031422 | 0.86 |
ENST00000249363.4
|
LRRC4
|
leucine rich repeat containing 4 |
| chr22_+_31496387 | 0.86 |
ENST00000443011.5
ENST00000400288.7 ENST00000400289.5 ENST00000444859.5 |
SFI1
|
SFI1 centrin binding protein |
| chr8_+_133191029 | 0.85 |
ENST00000250160.11
|
CCN4
|
cellular communication network factor 4 |
| chr17_+_77281429 | 0.83 |
ENST00000591198.5
ENST00000427177.6 |
SEPTIN9
|
septin 9 |
| chr14_-_91253925 | 0.83 |
ENST00000531499.2
|
GPR68
|
G protein-coupled receptor 68 |
| chr12_-_14961256 | 0.83 |
ENST00000541380.5
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr3_-_50503597 | 0.83 |
ENST00000266039.7
ENST00000424201.7 |
CACNA2D2
|
calcium voltage-gated channel auxiliary subunit alpha2delta 2 |
| chr3_-_52231190 | 0.82 |
ENST00000494383.1
|
ENSG00000173366.11
|
novel twinfilin, actin-binding protein, homolog 2 (Drosophila) (TWF2) and toll-like receptor 9 (TLR9) protein |
| chr19_+_41883173 | 0.81 |
ENST00000599846.5
ENST00000354532.8 ENST00000347545.8 |
ARHGEF1
|
Rho guanine nucleotide exchange factor 1 |
| chr10_+_11005301 | 0.81 |
ENST00000416382.6
ENST00000631460.1 ENST00000631816.1 |
CELF2
|
CUGBP Elav-like family member 2 |
| chr22_+_31944500 | 0.81 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein eta |
| chr18_+_3450036 | 0.81 |
ENST00000546979.5
ENST00000343820.10 ENST00000551402.1 ENST00000577543.5 |
TGIF1
|
TGFB induced factor homeobox 1 |
| chr9_-_112890868 | 0.81 |
ENST00000374228.5
|
SLC46A2
|
solute carrier family 46 member 2 |
| chr2_-_73284431 | 0.80 |
ENST00000521871.5
ENST00000520530.3 |
FBXO41
|
F-box protein 41 |
| chr5_-_177303675 | 0.80 |
ENST00000393611.6
ENST00000303270.6 ENST00000303251.11 |
RAB24
|
RAB24, member RAS oncogene family |
| chr17_+_7844915 | 0.80 |
ENST00000570632.1
|
KDM6B
|
lysine demethylase 6B |
| chr1_-_15585015 | 0.79 |
ENST00000375826.4
|
AGMAT
|
agmatinase |
| chr12_-_52601458 | 0.79 |
ENST00000537672.6
ENST00000293745.7 |
KRT72
|
keratin 72 |
| chr19_-_11578917 | 0.79 |
ENST00000649386.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr6_+_20403679 | 0.78 |
ENST00000535432.2
|
E2F3
|
E2F transcription factor 3 |
| chr4_-_77819615 | 0.78 |
ENST00000512485.6
|
CNOT6L
|
CCR4-NOT transcription complex subunit 6 like |
| chr7_-_87220520 | 0.78 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243 |
| chr22_+_31944527 | 0.78 |
ENST00000248975.6
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein eta |
| chr3_+_42502592 | 0.78 |
ENST00000438259.6
ENST00000543411.5 ENST00000439731.5 ENST00000325123.5 |
VIPR1
|
vasoactive intestinal peptide receptor 1 |
| chr7_-_27102669 | 0.78 |
ENST00000222718.7
|
HOXA2
|
homeobox A2 |
| chr4_+_26860809 | 0.77 |
ENST00000465503.6
ENST00000467087.7 |
STIM2
|
stromal interaction molecule 2 |
| chr16_-_71289367 | 0.77 |
ENST00000434935.7
ENST00000565850.1 ENST00000568910.1 |
CMTR2
|
cap methyltransferase 2 |
| chr2_+_202634960 | 0.77 |
ENST00000392238.3
|
FAM117B
|
family with sequence similarity 117 member B |
| chrX_-_154805516 | 0.76 |
ENST00000413259.7
|
MPP1
|
membrane palmitoylated protein 1 |
| chr19_+_7395166 | 0.76 |
ENST00000594665.2
ENST00000617428.4 |
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor 18 |
| chr7_-_65982205 | 0.75 |
ENST00000421103.5
ENST00000304895.9 |
GUSB
|
glucuronidase beta |
| chr9_+_87497852 | 0.75 |
ENST00000408954.8
|
DAPK1
|
death associated protein kinase 1 |
| chr7_-_25180278 | 0.75 |
ENST00000283905.8
ENST00000409280.5 ENST00000415598.5 |
C7orf31
|
chromosome 7 open reading frame 31 |
| chr9_-_120877167 | 0.74 |
ENST00000373896.8
ENST00000312189.10 |
PHF19
|
PHD finger protein 19 |
| chr8_-_58659534 | 0.74 |
ENST00000427130.6
|
NSMAF
|
neutral sphingomyelinase activation associated factor |
| chr16_+_30949054 | 0.74 |
ENST00000318663.5
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr19_+_7395112 | 0.74 |
ENST00000319670.14
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor 18 |
| chr14_+_22871732 | 0.74 |
ENST00000359591.9
|
LRP10
|
LDL receptor related protein 10 |
| chr4_+_77157189 | 0.73 |
ENST00000316355.10
ENST00000502280.5 |
CCNG2
|
cyclin G2 |
| chr10_+_26438317 | 0.73 |
ENST00000376236.9
|
APBB1IP
|
amyloid beta precursor protein binding family B member 1 interacting protein |
| chr11_+_117327829 | 0.73 |
ENST00000533153.5
ENST00000278935.8 ENST00000525416.5 |
CEP164
|
centrosomal protein 164 |
| chr17_+_48908461 | 0.73 |
ENST00000508468.2
|
UBE2Z
|
ubiquitin conjugating enzyme E2 Z |
| chr19_-_35900532 | 0.72 |
ENST00000396901.5
ENST00000641389.2 ENST00000585925.7 |
NFKBID
|
NFKB inhibitor delta |
| chr2_+_184598520 | 0.72 |
ENST00000302277.7
|
ZNF804A
|
zinc finger protein 804A |
| chr2_-_207166159 | 0.71 |
ENST00000426163.5
|
KLF7
|
Kruppel like factor 7 |
| chr3_+_9749940 | 0.71 |
ENST00000302003.11
ENST00000349503.9 ENST00000302036.11 ENST00000344629.12 ENST00000339511.9 |
OGG1
|
8-oxoguanine DNA glycosylase |
| chr16_+_81645352 | 0.70 |
ENST00000398040.8
|
CMIP
|
c-Maf inducing protein |
| chr4_+_26860778 | 0.70 |
ENST00000467011.6
|
STIM2
|
stromal interaction molecule 2 |
| chr19_-_19663664 | 0.70 |
ENST00000357324.11
|
ATP13A1
|
ATPase 13A1 |
| chr11_+_66070256 | 0.68 |
ENST00000320580.9
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chrX_+_30653359 | 0.68 |
ENST00000378943.7
ENST00000378946.7 ENST00000427190.6 |
GK
|
glycerol kinase |
| chr9_-_131276499 | 0.68 |
ENST00000372271.4
|
FAM78A
|
family with sequence similarity 78 member A |
| chr22_-_46537593 | 0.68 |
ENST00000262738.9
ENST00000674500.2 |
CELSR1
|
cadherin EGF LAG seven-pass G-type receptor 1 |
| chr18_+_3449620 | 0.68 |
ENST00000405385.7
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr22_+_44677044 | 0.68 |
ENST00000006251.11
|
PRR5
|
proline rich 5 |
| chr14_-_23010122 | 0.67 |
ENST00000397377.5
ENST00000397379.7 ENST00000341470.8 ENST00000555998.5 ENST00000299088.11 ENST00000397376.6 ENST00000553675.5 ENST00000553931.5 ENST00000555575.5 ENST00000553958.5 ENST00000555098.5 ENST00000556419.5 ENST00000553606.5 ENST00000554179.5 ENST00000397382.8 |
C14orf93
|
chromosome 14 open reading frame 93 |
| chr10_-_58267868 | 0.67 |
ENST00000373935.4
|
IPMK
|
inositol polyphosphate multikinase |
| chr16_-_71289609 | 0.67 |
ENST00000338099.9
ENST00000563876.1 |
CMTR2
|
cap methyltransferase 2 |
| chr3_+_23917170 | 0.67 |
ENST00000643707.1
|
RPL15
|
ribosomal protein L15 |
| chr16_-_28211476 | 0.66 |
ENST00000569951.1
ENST00000565698.5 |
XPO6
|
exportin 6 |
| chr1_+_154405326 | 0.66 |
ENST00000368485.8
|
IL6R
|
interleukin 6 receptor |
| chr12_+_121804689 | 0.66 |
ENST00000619791.1
ENST00000267197.9 |
SETD1B
|
SET domain containing 1B, histone lysine methyltransferase |
| chr12_-_6606427 | 0.66 |
ENST00000642879.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr9_+_113580912 | 0.65 |
ENST00000471324.6
|
RGS3
|
regulator of G protein signaling 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP2B
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.9 | 4.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.9 | 2.6 | GO:0006212 | uracil catabolic process(GO:0006212) beta-alanine biosynthetic process(GO:0019483) |
| 0.7 | 2.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.6 | 4.5 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.6 | 2.5 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.6 | 1.7 | GO:1901291 | negative regulation of double-strand break repair via single-strand annealing(GO:1901291) |
| 0.5 | 4.9 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.5 | 3.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.5 | 1.5 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.5 | 1.4 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.5 | 1.4 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.4 | 2.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.4 | 1.2 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.4 | 5.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.3 | 3.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 2.4 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.3 | 4.7 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 3.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.3 | 1.2 | GO:2000755 | regulation of phospholipid scramblase activity(GO:1900161) positive regulation of phospholipid scramblase activity(GO:1900163) regulation of glucosylceramide catabolic process(GO:2000752) positive regulation of glucosylceramide catabolic process(GO:2000753) regulation of sphingomyelin catabolic process(GO:2000754) positive regulation of sphingomyelin catabolic process(GO:2000755) |
| 0.3 | 0.8 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.3 | 1.8 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.2 | 1.2 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.2 | 1.9 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 0.9 | GO:2000595 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.2 | 0.9 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.2 | 2.3 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 0.8 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.2 | 1.9 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.2 | 0.7 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.2 | 7.9 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.2 | 0.5 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.2 | 1.3 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.2 | 0.5 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.6 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 1.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 3.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.3 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 1.7 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 0.5 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.8 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 1.8 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.5 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 1.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 1.0 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.1 | 2.8 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.5 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.5 | GO:2000607 | regulation of cell proliferation involved in mesonephros development(GO:2000606) negative regulation of cell proliferation involved in mesonephros development(GO:2000607) fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000699) glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000701) regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000702) negative regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000703) regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000733) negative regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000734) |
| 0.1 | 0.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.3 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.8 | GO:0061324 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.1 | 0.5 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.1 | 0.6 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.6 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.5 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 1.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 1.9 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.1 | 8.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.7 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.6 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.4 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.8 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.6 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.1 | 0.8 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 2.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.5 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.8 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 5.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 1.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.5 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.6 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 3.1 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 1.9 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.2 | GO:1902910 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.7 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.4 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 9.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 1.0 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 3.7 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.7 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 1.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.8 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.2 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.4 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 1.1 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.1 | 1.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 1.2 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.3 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 1.7 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.1 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.0 | 0.6 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.1 | GO:0018963 | phthalate metabolic process(GO:0018963) |
| 0.0 | 0.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 1.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.7 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 1.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 1.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 2.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.5 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.3 | GO:0061205 | alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.0 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.2 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.3 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 1.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.7 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.7 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.5 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 2.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 4.0 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.3 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 0.0 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.4 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.4 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.0 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 5.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 1.6 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.3 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0060947 | positive regulation of transcription via serum response element binding(GO:0010735) cardiac vascular smooth muscle cell differentiation(GO:0060947) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.8 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 1.4 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 1.8 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 1.6 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.3 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:1904903 | abscission(GO:0009838) ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.5 | GO:0051292 | nuclear pore complex assembly(GO:0051292) endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.3 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 2.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.3 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 | 0.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.9 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.3 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 2.9 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.8 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 0.7 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 9.3 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 1.2 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.6 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.4 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 1.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.3 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.7 | 6.8 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.4 | 4.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.4 | 1.5 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 1.4 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.2 | 0.6 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 1.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.6 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 4.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 1.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.3 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 1.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.7 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.5 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 13.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 2.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 2.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 7.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 6.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 2.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 0.3 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 1.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 1.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 1.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 2.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.6 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.6 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 2.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 3.3 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 3.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 7.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.5 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 2.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 3.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 1.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.6 | 1.7 | GO:0034039 | 8-oxo-7,8-dihydroguanine DNA N-glycosylase activity(GO:0034039) |
| 0.6 | 1.7 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 0.5 | 4.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.5 | 1.4 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.4 | 2.8 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.3 | 3.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 3.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 4.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 0.8 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.2 | 0.7 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.2 | 1.2 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.2 | 4.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 0.9 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.2 | 0.6 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.2 | 1.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 0.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 0.5 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 4.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 2.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 0.5 | GO:0052858 | peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) |
| 0.2 | 4.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 3.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 0.8 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.7 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.7 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.8 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.4 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.1 | 4.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 5.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 6.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.3 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.7 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.4 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.1 | 0.4 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.1 | 0.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.2 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.8 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.5 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 2.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.6 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.5 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.6 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 2.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 1.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.2 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.4 | GO:0005119 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 2.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 3.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 2.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.2 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 2.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 1.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 1.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 8.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 1.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 5.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.0 | 2.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 1.2 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 4.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.0 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 7.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 3.3 | GO:0017016 | Ras GTPase binding(GO:0017016) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 13.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 1.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 5.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 4.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 6.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 5.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.0 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 0.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 2.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 2.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 4.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 7.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 3.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 3.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.3 | 7.8 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 2.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.8 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.6 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 4.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 3.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 3.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 4.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 3.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 0.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 3.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 7.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.4 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 2.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 6.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 4.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 2.1 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.1 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.5 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.8 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |