Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
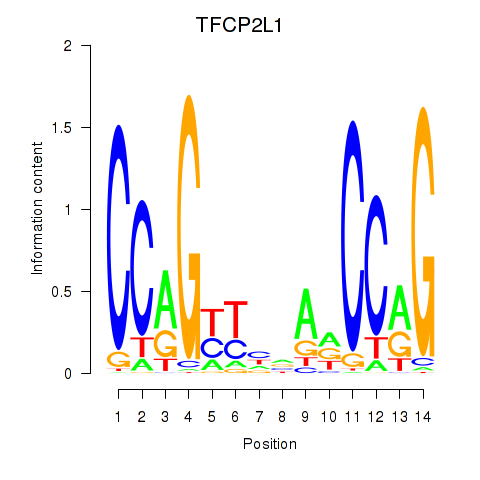
Results for TFCP2L1
Z-value: 1.41
Transcription factors associated with TFCP2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2L1
|
ENSG00000115112.8 | transcription factor CP2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2L1 | hg38_v1_chr2_-_121285194_121285209 | 0.18 | 3.3e-01 | Click! |
Activity profile of TFCP2L1 motif
Sorted Z-values of TFCP2L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_46413548 | 4.05 |
ENST00000307522.5
|
CCDC8
|
coiled-coil domain containing 8 |
| chr12_+_40692413 | 3.56 |
ENST00000551295.7
ENST00000547702.5 ENST00000551424.5 |
CNTN1
|
contactin 1 |
| chr1_-_153565535 | 2.93 |
ENST00000368707.5
|
S100A2
|
S100 calcium binding protein A2 |
| chr16_-_65121930 | 2.87 |
ENST00000566827.5
ENST00000394156.7 ENST00000268603.9 ENST00000562998.1 |
CDH11
|
cadherin 11 |
| chr11_-_119423162 | 2.84 |
ENST00000284240.10
ENST00000524970.5 |
THY1
|
Thy-1 cell surface antigen |
| chr15_-_51094897 | 2.75 |
ENST00000637513.2
|
TNFAIP8L3
|
TNF alpha induced protein 8 like 3 |
| chr1_-_225941383 | 2.71 |
ENST00000420304.6
|
LEFTY2
|
left-right determination factor 2 |
| chr1_-_225941212 | 2.64 |
ENST00000366820.10
|
LEFTY2
|
left-right determination factor 2 |
| chr1_-_204359885 | 2.59 |
ENST00000414478.1
ENST00000272203.8 |
PLEKHA6
|
pleckstrin homology domain containing A6 |
| chr15_+_74174403 | 2.51 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine rich repeat |
| chr2_-_55923775 | 2.25 |
ENST00000438672.5
ENST00000355426.8 ENST00000440439.5 ENST00000429909.5 ENST00000424207.5 ENST00000452337.5 ENST00000439193.5 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin extracellular matrix protein 1 |
| chr5_+_141475928 | 2.23 |
ENST00000611950.1
ENST00000308177.5 ENST00000617641.4 ENST00000621008.1 ENST00000617222.4 |
PCDHGC3
|
protocadherin gamma subfamily C, 3 |
| chr9_-_124507382 | 2.23 |
ENST00000373588.9
ENST00000620110.4 |
NR5A1
|
nuclear receptor subfamily 5 group A member 1 |
| chr6_-_31730198 | 2.13 |
ENST00000375787.6
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr14_+_58637934 | 2.04 |
ENST00000395153.8
|
DACT1
|
dishevelled binding antagonist of beta catenin 1 |
| chr9_-_127122623 | 2.03 |
ENST00000373417.1
ENST00000373425.8 |
ANGPTL2
|
angiopoietin like 2 |
| chr11_-_119423193 | 2.02 |
ENST00000580275.5
|
THY1
|
Thy-1 cell surface antigen |
| chr1_+_26863140 | 2.02 |
ENST00000339276.6
|
SFN
|
stratifin |
| chr17_+_42851167 | 2.00 |
ENST00000613571.1
ENST00000308423.7 |
AOC3
|
amine oxidase copper containing 3 |
| chr17_+_7888783 | 1.92 |
ENST00000330494.12
ENST00000358181.8 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr1_+_204870831 | 1.89 |
ENST00000404076.5
ENST00000539706.6 |
NFASC
|
neurofascin |
| chr1_-_1421302 | 1.87 |
ENST00000520296.5
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr1_-_155990062 | 1.80 |
ENST00000462460.6
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr20_+_56629296 | 1.80 |
ENST00000201031.3
|
TFAP2C
|
transcription factor AP-2 gamma |
| chr19_-_12140315 | 1.79 |
ENST00000418866.1
ENST00000600335.5 ENST00000334213.10 |
ZNF20
|
zinc finger protein 20 |
| chr1_-_1421248 | 1.78 |
ENST00000442470.1
ENST00000537107.6 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr10_-_98011320 | 1.75 |
ENST00000309155.3
|
CRTAC1
|
cartilage acidic protein 1 |
| chr1_-_45623967 | 1.74 |
ENST00000445048.2
ENST00000421127.6 ENST00000528266.6 |
CCDC17
|
coiled-coil domain containing 17 |
| chr20_+_36154630 | 1.71 |
ENST00000338074.7
ENST00000636016.2 ENST00000373945.5 |
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr14_-_68937942 | 1.68 |
ENST00000684182.1
|
ACTN1
|
actinin alpha 1 |
| chr19_+_47713412 | 1.67 |
ENST00000538399.1
ENST00000263277.8 |
EHD2
|
EH domain containing 2 |
| chr17_-_28711660 | 1.64 |
ENST00000439862.7
ENST00000581289.1 ENST00000301039.6 ENST00000682792.1 |
PROCA1
|
protein interacting with cyclin A1 |
| chr3_-_123692347 | 1.63 |
ENST00000508240.1
|
MYLK
|
myosin light chain kinase |
| chr16_+_84294853 | 1.58 |
ENST00000219454.10
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr16_+_84294823 | 1.56 |
ENST00000568638.1
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr6_-_31729260 | 1.56 |
ENST00000375789.7
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr14_-_106803221 | 1.55 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr1_-_111200633 | 1.51 |
ENST00000357640.9
|
DENND2D
|
DENN domain containing 2D |
| chr3_+_71753834 | 1.51 |
ENST00000304411.3
|
GPR27
|
G protein-coupled receptor 27 |
| chr5_+_122311740 | 1.47 |
ENST00000506272.5
ENST00000508681.5 ENST00000509154.6 |
SNCAIP
|
synuclein alpha interacting protein |
| chr5_+_52989314 | 1.47 |
ENST00000296585.10
|
ITGA2
|
integrin subunit alpha 2 |
| chr14_+_96204679 | 1.46 |
ENST00000542454.2
ENST00000539359.1 ENST00000554311.2 ENST00000553811.1 |
BDKRB2
ENSG00000258691.1
|
bradykinin receptor B2 novel protein |
| chr21_-_30216047 | 1.45 |
ENST00000399899.2
|
CLDN8
|
claudin 8 |
| chr1_+_27342014 | 1.45 |
ENST00000618673.4
ENST00000318074.9 ENST00000616558.5 |
SYTL1
|
synaptotagmin like 1 |
| chr1_-_182604379 | 1.45 |
ENST00000367558.6
|
RGS16
|
regulator of G protein signaling 16 |
| chr5_+_177086867 | 1.44 |
ENST00000503708.5
ENST00000393648.6 ENST00000514472.1 ENST00000502906.5 ENST00000292408.9 ENST00000510911.5 |
FGFR4
|
fibroblast growth factor receptor 4 |
| chr12_+_53097656 | 1.41 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr8_-_6877928 | 1.41 |
ENST00000297439.4
|
DEFB1
|
defensin beta 1 |
| chr19_-_20661563 | 1.38 |
ENST00000601440.6
ENST00000291750.6 ENST00000595094.1 |
ZNF626
ENSG00000269110.1
|
zinc finger protein 626 novel transcript |
| chr4_+_84583037 | 1.35 |
ENST00000295887.6
|
CDS1
|
CDP-diacylglycerol synthase 1 |
| chr14_-_106154113 | 1.35 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr1_-_58784035 | 1.32 |
ENST00000371222.4
|
JUN
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr4_-_25863537 | 1.32 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr10_-_119536533 | 1.31 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr1_-_207032749 | 1.29 |
ENST00000359470.6
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr7_+_30921430 | 1.28 |
ENST00000409899.5
ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr11_+_62613236 | 1.28 |
ENST00000278833.4
|
ROM1
|
retinal outer segment membrane protein 1 |
| chr11_-_62612725 | 1.27 |
ENST00000419857.1
ENST00000394773.7 |
EML3
|
EMAP like 3 |
| chr5_-_180591488 | 1.26 |
ENST00000292641.4
|
SCGB3A1
|
secretoglobin family 3A member 1 |
| chr12_+_93574965 | 1.25 |
ENST00000551883.1
ENST00000549510.1 |
SOCS2
|
suppressor of cytokine signaling 2 |
| chr15_+_73926443 | 1.24 |
ENST00000261921.8
|
LOXL1
|
lysyl oxidase like 1 |
| chr7_+_64313943 | 1.23 |
ENST00000423484.3
|
ZNF736
|
zinc finger protein 736 |
| chr6_-_32763522 | 1.20 |
ENST00000435145.6
ENST00000437316.7 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr8_-_53842899 | 1.20 |
ENST00000524234.1
ENST00000521275.5 ENST00000396774.6 |
ATP6V1H
|
ATPase H+ transporting V1 subunit H |
| chr2_+_90234809 | 1.19 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr1_+_151540299 | 1.19 |
ENST00000392712.7
ENST00000368848.6 ENST00000368849.8 ENST00000353024.4 |
TUFT1
|
tuftelin 1 |
| chr8_-_27992663 | 1.19 |
ENST00000380385.6
ENST00000354914.8 |
SCARA5
|
scavenger receptor class A member 5 |
| chrX_+_134237047 | 1.18 |
ENST00000370809.4
ENST00000517294.5 |
CCDC160
|
coiled-coil domain containing 160 |
| chr6_-_31582415 | 1.15 |
ENST00000429299.3
ENST00000446745.2 |
LTB
|
lymphotoxin beta |
| chr2_-_168247569 | 1.15 |
ENST00000355999.5
|
STK39
|
serine/threonine kinase 39 |
| chr2_-_72147819 | 1.12 |
ENST00000001146.7
ENST00000546307.5 ENST00000474509.1 |
CYP26B1
|
cytochrome P450 family 26 subfamily B member 1 |
| chr19_-_20565769 | 1.12 |
ENST00000427401.9
|
ZNF737
|
zinc finger protein 737 |
| chr17_+_7654096 | 1.12 |
ENST00000577113.1
|
ATP1B2
|
ATPase Na+/K+ transporting subunit beta 2 |
| chr20_+_9514562 | 1.12 |
ENST00000246070.3
|
LAMP5
|
lysosomal associated membrane protein family member 5 |
| chr12_+_55931148 | 1.10 |
ENST00000549629.5
ENST00000555218.5 ENST00000331886.10 |
DGKA
|
diacylglycerol kinase alpha |
| chr7_-_128405930 | 1.10 |
ENST00000470772.5
ENST00000480861.5 ENST00000496200.5 |
IMPDH1
|
inosine monophosphate dehydrogenase 1 |
| chr13_-_36131286 | 1.09 |
ENST00000255448.8
ENST00000379892.4 |
DCLK1
|
doublecortin like kinase 1 |
| chr1_-_161038907 | 1.09 |
ENST00000318289.14
ENST00000368023.7 ENST00000423014.3 ENST00000368024.5 |
TSTD1
|
thiosulfate sulfurtransferase like domain containing 1 |
| chr14_+_58638023 | 1.08 |
ENST00000335867.4
|
DACT1
|
dishevelled binding antagonist of beta catenin 1 |
| chr19_-_20661507 | 1.05 |
ENST00000612591.4
ENST00000595405.1 |
ZNF626
|
zinc finger protein 626 |
| chrX_+_70455093 | 1.04 |
ENST00000542398.1
|
DLG3
|
discs large MAGUK scaffold protein 3 |
| chr2_+_158968608 | 1.03 |
ENST00000263635.8
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr8_-_97277890 | 1.03 |
ENST00000322128.5
|
TSPYL5
|
TSPY like 5 |
| chr11_-_128867364 | 1.02 |
ENST00000440599.6
ENST00000324036.7 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr17_-_19716604 | 1.02 |
ENST00000325411.9
ENST00000350657.9 |
SLC47A2
|
solute carrier family 47 member 2 |
| chr12_+_48814451 | 1.01 |
ENST00000540990.5
|
CACNB3
|
calcium voltage-gated channel auxiliary subunit beta 3 |
| chr12_+_52079700 | 1.00 |
ENST00000546390.2
|
SMIM41
|
small integral membrane protein 41 |
| chr17_-_15263162 | 0.99 |
ENST00000674673.1
ENST00000675950.1 |
PMP22
|
peripheral myelin protein 22 |
| chr11_-_128867268 | 0.99 |
ENST00000392665.6
ENST00000392666.6 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr7_-_100158679 | 0.97 |
ENST00000456769.5
ENST00000316937.8 |
TRAPPC14
|
trafficking protein particle complex 14 |
| chr4_-_163332589 | 0.96 |
ENST00000296533.3
ENST00000509586.5 ENST00000504391.5 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chr17_-_35448742 | 0.95 |
ENST00000534689.5
ENST00000532210.5 ENST00000285013.11 ENST00000526861.5 ENST00000531588.1 |
SLFN13
|
schlafen family member 13 |
| chr10_+_100347225 | 0.95 |
ENST00000370355.3
|
SCD
|
stearoyl-CoA desaturase |
| chr2_-_31138429 | 0.93 |
ENST00000349752.10
|
GALNT14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr19_-_11738882 | 0.92 |
ENST00000586121.1
ENST00000431998.1 ENST00000341191.11 ENST00000440527.1 |
ZNF823
|
zinc finger protein 823 |
| chr17_+_77376083 | 0.92 |
ENST00000427674.6
|
SEPTIN9
|
septin 9 |
| chr12_-_120369156 | 0.92 |
ENST00000257552.7
|
MSI1
|
musashi RNA binding protein 1 |
| chr19_-_12294819 | 0.90 |
ENST00000355684.6
ENST00000356109.10 |
ZNF44
|
zinc finger protein 44 |
| chr7_+_129188622 | 0.90 |
ENST00000249373.8
|
SMO
|
smoothened, frizzled class receptor |
| chr19_-_19821704 | 0.89 |
ENST00000443905.6
ENST00000590766.5 ENST00000587452.5 ENST00000545006.1 ENST00000590319.1 ENST00000587461.5 ENST00000450683.6 ENST00000540806.7 ENST00000590274.1 |
ZNF506
ENSG00000267581.1
|
zinc finger protein 506 novel transcript |
| chr6_-_32666648 | 0.89 |
ENST00000399082.7
ENST00000399079.7 ENST00000374943.8 ENST00000434651.6 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr11_-_65557740 | 0.89 |
ENST00000526927.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr13_-_36131352 | 0.88 |
ENST00000360631.8
|
DCLK1
|
doublecortin like kinase 1 |
| chr16_-_2340703 | 0.87 |
ENST00000301732.10
ENST00000382381.7 |
ABCA3
|
ATP binding cassette subfamily A member 3 |
| chr4_-_173334385 | 0.87 |
ENST00000446922.6
|
HMGB2
|
high mobility group box 2 |
| chr7_+_66205325 | 0.87 |
ENST00000304842.6
ENST00000649664.1 |
TPST1
|
tyrosylprotein sulfotransferase 1 |
| chr11_+_95089804 | 0.87 |
ENST00000278505.5
|
ENDOD1
|
endonuclease domain containing 1 |
| chr1_-_43285559 | 0.86 |
ENST00000523677.6
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr14_-_68979251 | 0.85 |
ENST00000438964.6
ENST00000679147.1 |
ACTN1
|
actinin alpha 1 |
| chr17_+_64506953 | 0.84 |
ENST00000580188.1
ENST00000556440.7 ENST00000581056.5 |
CEP95
|
centrosomal protein 95 |
| chr1_+_205256189 | 0.84 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr7_+_64903247 | 0.83 |
ENST00000476120.1
|
ZNF273
|
zinc finger protein 273 |
| chr3_-_64445396 | 0.82 |
ENST00000295902.11
|
PRICKLE2
|
prickle planar cell polarity protein 2 |
| chr12_+_119178920 | 0.82 |
ENST00000281938.7
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr2_-_20012661 | 0.81 |
ENST00000421259.2
ENST00000407540.8 |
MATN3
|
matrilin 3 |
| chr12_-_53232182 | 0.81 |
ENST00000425354.7
ENST00000546717.1 ENST00000394426.5 |
RARG
|
retinoic acid receptor gamma |
| chr1_+_54641754 | 0.80 |
ENST00000339553.9
ENST00000421030.7 |
MROH7
|
maestro heat like repeat family member 7 |
| chr12_+_41188301 | 0.80 |
ENST00000402685.7
|
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr6_-_52995170 | 0.79 |
ENST00000370959.1
ENST00000370963.9 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr16_+_31355165 | 0.79 |
ENST00000562918.5
ENST00000268296.9 |
ITGAX
|
integrin subunit alpha X |
| chr2_-_136118142 | 0.79 |
ENST00000241393.4
|
CXCR4
|
C-X-C motif chemokine receptor 4 |
| chr17_+_64507029 | 0.79 |
ENST00000553412.5
|
CEP95
|
centrosomal protein 95 |
| chr7_-_65006678 | 0.79 |
ENST00000394323.3
|
ERV3-1
|
endogenous retrovirus group 3 member 1, envelope |
| chr20_+_346769 | 0.78 |
ENST00000609179.5
|
NRSN2
|
neurensin 2 |
| chr1_+_116111395 | 0.78 |
ENST00000684484.1
ENST00000369500.4 |
MAB21L3
|
mab-21 like 3 |
| chr15_+_100877714 | 0.78 |
ENST00000561338.5
|
ALDH1A3
|
aldehyde dehydrogenase 1 family member A3 |
| chr20_-_54173976 | 0.77 |
ENST00000216862.8
|
CYP24A1
|
cytochrome P450 family 24 subfamily A member 1 |
| chr14_-_106622837 | 0.77 |
ENST00000390628.3
|
IGHV1-58
|
immunoglobulin heavy variable 1-58 |
| chr8_+_54457927 | 0.77 |
ENST00000297316.5
|
SOX17
|
SRY-box transcription factor 17 |
| chr14_-_68979076 | 0.75 |
ENST00000538545.6
ENST00000684639.1 |
ACTN1
|
actinin alpha 1 |
| chr12_+_119178953 | 0.75 |
ENST00000674542.1
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr4_-_82430192 | 0.74 |
ENST00000621267.4
ENST00000614627.4 |
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D like |
| chr19_-_20565746 | 0.73 |
ENST00000594419.1
|
ZNF737
|
zinc finger protein 737 |
| chr7_-_128031422 | 0.73 |
ENST00000249363.4
|
LRRC4
|
leucine rich repeat containing 4 |
| chr19_-_1822038 | 0.73 |
ENST00000643515.1
|
REXO1
|
RNA exonuclease 1 homolog |
| chr7_+_6615576 | 0.72 |
ENST00000457543.4
|
ZNF853
|
zinc finger protein 853 |
| chr17_-_76027296 | 0.72 |
ENST00000301607.8
|
EVPL
|
envoplakin |
| chr10_-_126388455 | 0.72 |
ENST00000368679.8
ENST00000368676.8 ENST00000448723.2 |
ADAM12
|
ADAM metallopeptidase domain 12 |
| chr18_-_48409292 | 0.71 |
ENST00000589194.5
ENST00000591279.5 ENST00000590855.5 ENST00000587107.5 ENST00000588970.5 ENST00000586525.5 ENST00000592387.5 ENST00000590800.6 |
ZBTB7C
|
zinc finger and BTB domain containing 7C |
| chrX_+_151397163 | 0.71 |
ENST00000330374.7
|
VMA21
|
vacuolar ATPase assembly factor VMA21 |
| chr14_-_106130061 | 0.71 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr20_-_33686371 | 0.71 |
ENST00000343380.6
|
E2F1
|
E2F transcription factor 1 |
| chr8_+_73008850 | 0.70 |
ENST00000679115.1
ENST00000517390.2 ENST00000678860.1 ENST00000678518.1 ENST00000276602.10 ENST00000276603.10 ENST00000518874.6 |
TERF1
|
telomeric repeat binding factor 1 |
| chr14_-_68979314 | 0.70 |
ENST00000684713.1
ENST00000683198.1 ENST00000684598.1 ENST00000682331.1 ENST00000682291.1 ENST00000683342.1 |
ACTN1
|
actinin alpha 1 |
| chr2_+_240605417 | 0.70 |
ENST00000319838.10
ENST00000403859.1 |
GPR35
|
G protein-coupled receptor 35 |
| chr7_-_1028311 | 0.70 |
ENST00000412051.5
|
C7orf50
|
chromosome 7 open reading frame 50 |
| chr17_+_16415553 | 0.70 |
ENST00000338560.12
|
TRPV2
|
transient receptor potential cation channel subfamily V member 2 |
| chr3_-_50340804 | 0.69 |
ENST00000359365.9
ENST00000357043.6 |
RASSF1
|
Ras association domain family member 1 |
| chr2_+_188291911 | 0.69 |
ENST00000410051.5
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr12_+_57772587 | 0.69 |
ENST00000300209.13
|
EEF1AKMT3
|
EEF1A lysine methyltransferase 3 |
| chr17_-_19716573 | 0.69 |
ENST00000433844.4
|
SLC47A2
|
solute carrier family 47 member 2 |
| chr19_-_12052954 | 0.68 |
ENST00000547628.2
|
ZNF878
|
zinc finger protein 878 |
| chr9_+_113349514 | 0.68 |
ENST00000374183.5
|
BSPRY
|
B-box and SPRY domain containing |
| chr2_+_9206762 | 0.68 |
ENST00000315273.4
ENST00000281419.8 |
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr22_+_31093358 | 0.67 |
ENST00000404574.5
|
SMTN
|
smoothelin |
| chr19_+_12064720 | 0.67 |
ENST00000550826.1
ENST00000439326.8 |
ZNF844
|
zinc finger protein 844 |
| chr12_-_120904337 | 0.65 |
ENST00000353487.7
|
SPPL3
|
signal peptide peptidase like 3 |
| chr17_+_42844573 | 0.64 |
ENST00000253799.8
ENST00000452774.2 |
AOC2
|
amine oxidase copper containing 2 |
| chr11_+_78188797 | 0.64 |
ENST00000530267.5
|
USP35
|
ubiquitin specific peptidase 35 |
| chr7_+_64045420 | 0.64 |
ENST00000647787.1
ENST00000456806.3 |
ENSG00000285544.1
ZNF727
|
novel transcript zinc finger protein 727 |
| chr22_+_20131781 | 0.64 |
ENST00000334554.12
ENST00000320602.11 ENST00000405930.3 |
ZDHHC8
|
zinc finger DHHC-type palmitoyltransferase 8 |
| chr2_+_231781669 | 0.63 |
ENST00000410024.5
ENST00000611614.4 ENST00000409295.5 ENST00000409091.5 |
COPS7B
|
COP9 signalosome subunit 7B |
| chr14_-_68979274 | 0.62 |
ENST00000394419.9
|
ACTN1
|
actinin alpha 1 |
| chr5_+_72179622 | 0.62 |
ENST00000504492.1
|
MAP1B
|
microtubule associated protein 1B |
| chr19_-_12484773 | 0.61 |
ENST00000397732.8
|
ZNF709
|
zinc finger protein 709 |
| chr22_+_22734577 | 0.61 |
ENST00000390310.3
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr1_+_54641806 | 0.61 |
ENST00000409996.5
|
MROH7
|
maestro heat like repeat family member 7 |
| chrX_-_129523436 | 0.61 |
ENST00000371123.5
ENST00000371121.5 ENST00000371122.8 |
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr6_-_32763466 | 0.61 |
ENST00000427449.1
ENST00000411527.5 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr10_+_79068955 | 0.60 |
ENST00000334512.10
|
ZMIZ1
|
zinc finger MIZ-type containing 1 |
| chr3_-_50567646 | 0.60 |
ENST00000426034.5
ENST00000441239.5 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr16_+_31355215 | 0.60 |
ENST00000562522.2
|
ITGAX
|
integrin subunit alpha X |
| chr1_-_161550591 | 0.60 |
ENST00000367967.7
ENST00000436743.6 ENST00000442336.1 |
FCGR3A
|
Fc fragment of IgG receptor IIIa |
| chr2_+_71068636 | 0.58 |
ENST00000244204.11
ENST00000533981.5 |
NAGK
|
N-acetylglucosamine kinase |
| chr12_-_121274510 | 0.58 |
ENST00000392474.6
|
CAMKK2
|
calcium/calmodulin dependent protein kinase kinase 2 |
| chr22_-_22508702 | 0.57 |
ENST00000626650.3
|
ZNF280B
|
zinc finger protein 280B |
| chr12_+_57772648 | 0.57 |
ENST00000333012.5
|
EEF1AKMT3
|
EEF1A lysine methyltransferase 3 |
| chr11_+_78188871 | 0.57 |
ENST00000528910.5
ENST00000529308.6 |
USP35
|
ubiquitin specific peptidase 35 |
| chr4_-_173334249 | 0.55 |
ENST00000506267.1
ENST00000296503.10 |
HMGB2
|
high mobility group box 2 |
| chr5_-_149379286 | 0.54 |
ENST00000261796.4
|
IL17B
|
interleukin 17B |
| chr1_-_155978524 | 0.54 |
ENST00000361247.9
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr19_+_41219235 | 0.53 |
ENST00000359092.7
|
AXL
|
AXL receptor tyrosine kinase |
| chr5_-_140043069 | 0.53 |
ENST00000289409.8
ENST00000358522.7 ENST00000289422.11 ENST00000541337.5 ENST00000361474.6 |
NRG2
|
neuregulin 2 |
| chr3_-_50340933 | 0.53 |
ENST00000616212.4
|
RASSF1
|
Ras association domain family member 1 |
| chr7_+_32957385 | 0.52 |
ENST00000538336.5
ENST00000242209.9 |
FKBP9
|
FKBP prolyl isomerase 9 |
| chr19_+_11965029 | 0.52 |
ENST00000592625.5
ENST00000358987.8 ENST00000586494.1 ENST00000343949.9 |
ZNF763
|
zinc finger protein 763 |
| chr19_+_11766989 | 0.51 |
ENST00000357901.5
|
ZNF441
|
zinc finger protein 441 |
| chr11_+_96389985 | 0.51 |
ENST00000332349.5
|
JRKL
|
JRK like |
| chr2_-_19990058 | 0.51 |
ENST00000281405.9
ENST00000345530.8 |
WDR35
|
WD repeat domain 35 |
| chr16_+_67528799 | 0.51 |
ENST00000379312.7
ENST00000042381.9 ENST00000540839.7 |
RIPOR1
|
RHO family interacting cell polarization regulator 1 |
| chr4_-_121823843 | 0.50 |
ENST00000274026.10
|
CCNA2
|
cyclin A2 |
| chr1_+_26545833 | 0.49 |
ENST00000531382.5
|
RPS6KA1
|
ribosomal protein S6 kinase A1 |
| chr10_+_100745570 | 0.49 |
ENST00000427256.6
ENST00000355243.8 |
PAX2
|
paired box 2 |
| chr4_-_82430399 | 0.48 |
ENST00000630827.1
ENST00000295470.10 |
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D like |
| chr19_+_20077967 | 0.48 |
ENST00000418063.3
|
ZNF90
|
zinc finger protein 90 |
| chr1_+_1425854 | 0.48 |
ENST00000378821.4
|
TMEM88B
|
transmembrane protein 88B |
| chr2_+_71068603 | 0.48 |
ENST00000443938.6
|
NAGK
|
N-acetylglucosamine kinase |
| chr3_+_158110363 | 0.48 |
ENST00000683137.1
|
RSRC1
|
arginine and serine rich coiled-coil 1 |
| chr17_+_44187210 | 0.48 |
ENST00000589785.1
ENST00000592825.1 ENST00000589184.5 |
TMUB2
|
transmembrane and ubiquitin like domain containing 2 |
| chr15_+_43594027 | 0.48 |
ENST00000453733.5
ENST00000441322.6 ENST00000627381.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2L1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.0 | 3.1 | GO:1904864 | regulation of beta-catenin-TCF complex assembly(GO:1904863) negative regulation of beta-catenin-TCF complex assembly(GO:1904864) |
| 0.5 | 1.5 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.5 | 2.4 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.4 | 1.3 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.4 | 1.1 | GO:2001035 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.3 | 2.0 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.3 | 1.3 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.3 | 2.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 1.2 | GO:0018277 | protein deamination(GO:0018277) |
| 0.3 | 1.5 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.3 | 2.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.3 | 1.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.3 | 1.1 | GO:0021634 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.3 | 1.4 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.3 | 3.3 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.3 | 0.8 | GO:0060796 | inner cell mass cellular morphogenesis(GO:0001828) regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) cardiac cell fate determination(GO:0060913) |
| 0.2 | 1.7 | GO:0046618 | drug export(GO:0046618) |
| 0.2 | 0.7 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.2 | 1.6 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.2 | 0.9 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 1.4 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.2 | 1.7 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.2 | 1.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 1.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 0.9 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) pancreas morphogenesis(GO:0061113) |
| 0.2 | 0.9 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.2 | 1.1 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.2 | 0.5 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.2 | 3.7 | GO:0000052 | citrulline metabolic process(GO:0000052) arginine catabolic process(GO:0006527) |
| 0.2 | 5.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 1.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 0.8 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.6 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.4 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.1 | 0.7 | GO:0032214 | regulation of telomere maintenance via semi-conservative replication(GO:0032213) negative regulation of telomere maintenance via semi-conservative replication(GO:0032214) |
| 0.1 | 0.9 | GO:2000669 | positive regulation of pinocytosis(GO:0048549) negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.1 | 1.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 1.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.2 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.1 | 2.9 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 0.4 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.6 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.5 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 1.4 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 1.8 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.8 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.6 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.4 | GO:0060529 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.1 | 0.3 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.3 | GO:1905071 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.1 | 0.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 1.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.2 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:1904828 | regulation of phenotypic switching by transcription from RNA polymerase II promoter(GO:0100057) regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.0 | 0.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 1.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.4 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.3 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 2.8 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.7 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 1.2 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 2.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.3 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 2.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 3.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.6 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 2.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 2.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.5 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.7 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 1.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 1.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 4.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.5 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 1.0 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 1.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 1.0 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 1.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.4 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.3 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 3.3 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 0.0 | 0.8 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 1.3 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 3.6 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.6 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.8 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 3.0 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0060733 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.5 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.0 | 1.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 1.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 3.4 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 1.9 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 1.0 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.0 | 0.3 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 2.0 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 1.2 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.7 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 1.7 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.5 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.7 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.9 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.0 | 2.9 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.7 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 1.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.1 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.0 | GO:1990393 | 3M complex(GO:1990393) |
| 0.4 | 1.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.4 | 1.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 1.4 | GO:1990742 | microvesicle(GO:1990742) |
| 0.2 | 1.5 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.2 | 0.7 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 1.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 1.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 0.8 | GO:0019031 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.2 | 0.9 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.2 | 4.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.4 | GO:1990917 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.1 | 4.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 2.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 3.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.6 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 1.4 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.2 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.8 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 7.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 3.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.7 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.5 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.9 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 0.2 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 1.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 1.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 2.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 1.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 3.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.5 | 2.0 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.5 | 3.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.5 | 2.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 4.9 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.4 | 1.3 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.4 | 1.7 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.4 | 1.4 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.4 | 1.4 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.3 | 1.7 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.3 | 1.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.2 | 1.0 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.2 | 3.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 0.9 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 1.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 1.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 2.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 2.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 1.5 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 1.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 2.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 0.8 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.4 | GO:0002135 | CTP binding(GO:0002135) sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 1.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.9 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 1.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 1.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 5.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.4 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 1.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 1.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.5 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 3.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 1.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 1.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 3.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 1.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.2 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 1.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.3 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 2.4 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 1.3 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 6.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 4.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 6.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 4.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 3.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 3.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 2.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 4.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 5.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 5.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 3.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.4 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 1.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 1.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 2.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 2.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.8 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 7.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |