Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
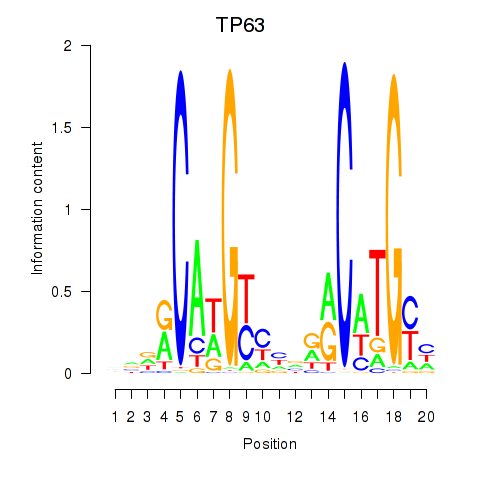
Results for TP63
Z-value: 1.76
Transcription factors associated with TP63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP63
|
ENSG00000073282.14 | tumor protein p63 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP63 | hg38_v1_chr3_+_189789672_189789681 | 0.39 | 2.7e-02 | Click! |
Activity profile of TP63 motif
Sorted Z-values of TP63 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_119431846 | 5.64 |
ENST00000306406.5
|
TMEM37
|
transmembrane protein 37 |
| chr1_-_153615858 | 4.38 |
ENST00000476873.5
|
S100A14
|
S100 calcium binding protein A14 |
| chr1_-_47190013 | 4.15 |
ENST00000294338.7
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr19_+_18386150 | 3.94 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr6_-_46954922 | 3.92 |
ENST00000265417.7
|
ADGRF5
|
adhesion G protein-coupled receptor F5 |
| chr7_+_80638510 | 3.82 |
ENST00000433696.6
ENST00000538969.5 ENST00000544133.5 |
CD36
|
CD36 molecule |
| chr11_+_18266254 | 3.74 |
ENST00000532858.5
ENST00000649195.1 ENST00000356524.9 ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr10_-_88991282 | 3.61 |
ENST00000458159.5
ENST00000415557.1 |
ACTA2
|
actin alpha 2, smooth muscle |
| chr11_-_111911759 | 3.61 |
ENST00000650687.2
|
CRYAB
|
crystallin alpha B |
| chrX_+_2752024 | 3.52 |
ENST00000644266.2
ENST00000419513.7 ENST00000509484.3 ENST00000381174.10 |
XG
|
Xg glycoprotein (Xg blood group) |
| chr13_-_77918820 | 3.51 |
ENST00000646607.2
|
EDNRB
|
endothelin receptor type B |
| chr2_+_216659600 | 3.36 |
ENST00000456764.1
|
IGFBP2
|
insulin like growth factor binding protein 2 |
| chr1_-_209651291 | 3.30 |
ENST00000391911.5
ENST00000415782.1 |
LAMB3
|
laminin subunit beta 3 |
| chr7_+_80638662 | 3.20 |
ENST00000394788.7
|
CD36
|
CD36 molecule |
| chr13_-_77918788 | 3.19 |
ENST00000626030.1
|
EDNRB
|
endothelin receptor type B |
| chr7_+_80638633 | 3.13 |
ENST00000447544.7
ENST00000482059.6 |
CD36
|
CD36 molecule |
| chr14_+_51489112 | 3.13 |
ENST00000356218.8
|
FRMD6
|
FERM domain containing 6 |
| chr15_+_74174403 | 3.08 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine rich repeat |
| chr4_+_25648011 | 3.03 |
ENST00000645788.1
|
SLC34A2
|
solute carrier family 34 member 2 |
| chr2_-_150487658 | 2.92 |
ENST00000375734.6
ENST00000263895.9 ENST00000454202.5 |
RND3
|
Rho family GTPase 3 |
| chr12_+_119178920 | 2.89 |
ENST00000281938.7
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr12_+_119178953 | 2.86 |
ENST00000674542.1
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr15_+_40844018 | 2.86 |
ENST00000344051.8
ENST00000562057.6 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr6_+_121435595 | 2.82 |
ENST00000649003.1
ENST00000282561.4 |
GJA1
|
gap junction protein alpha 1 |
| chr19_-_4518465 | 2.81 |
ENST00000633942.1
|
PLIN4
|
perilipin 4 |
| chr2_+_233691607 | 2.75 |
ENST00000373424.5
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chr9_+_35906179 | 2.75 |
ENST00000354323.3
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr6_-_42142604 | 2.73 |
ENST00000356542.5
ENST00000341865.9 |
C6orf132
|
chromosome 6 open reading frame 132 |
| chr15_-_88913362 | 2.71 |
ENST00000558029.5
ENST00000268150.13 ENST00000542878.5 ENST00000268151.11 ENST00000566497.5 |
MFGE8
|
milk fat globule EGF and factor V/VIII domain containing |
| chr20_-_54173976 | 2.64 |
ENST00000216862.8
|
CYP24A1
|
cytochrome P450 family 24 subfamily A member 1 |
| chr1_-_12618198 | 2.58 |
ENST00000616661.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr10_-_86969178 | 2.57 |
ENST00000440490.1
ENST00000609457.5 |
ADIRF-AS1
MMRN2
|
ADIRF antisense RNA 1 multimerin 2 |
| chr11_-_120138104 | 2.49 |
ENST00000341846.10
|
TRIM29
|
tripartite motif containing 29 |
| chr11_+_76782250 | 2.48 |
ENST00000533752.1
ENST00000612930.1 |
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr10_-_86969442 | 2.44 |
ENST00000474994.2
|
MMRN2
|
multimerin 2 |
| chr2_-_164842011 | 2.40 |
ENST00000409184.8
ENST00000456693.5 |
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr11_+_111912725 | 2.30 |
ENST00000304298.4
|
HSPB2
|
heat shock protein family B (small) member 2 |
| chr3_-_50303565 | 2.29 |
ENST00000266031.8
ENST00000395143.6 ENST00000457214.6 ENST00000447605.2 ENST00000395144.7 ENST00000418723.1 |
HYAL1
|
hyaluronidase 1 |
| chr12_-_88580459 | 2.27 |
ENST00000552044.1
ENST00000644744.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chrX_-_154374623 | 2.23 |
ENST00000369850.10
|
FLNA
|
filamin A |
| chr16_-_67190099 | 2.21 |
ENST00000314586.11
ENST00000563889.1 ENST00000564418.1 ENST00000545725.6 |
EXOC3L1
|
exocyst complex component 3 like 1 |
| chr3_+_111911604 | 2.18 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology like domain family B member 2 |
| chr3_+_132597260 | 2.17 |
ENST00000249887.3
|
ACKR4
|
atypical chemokine receptor 4 |
| chr2_-_164842140 | 2.16 |
ENST00000496396.1
ENST00000629362.2 ENST00000445474.2 ENST00000483743.6 |
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr19_-_5293232 | 2.15 |
ENST00000591760.5
|
PTPRS
|
protein tyrosine phosphatase receptor type S |
| chr19_-_17264718 | 2.14 |
ENST00000431146.6
ENST00000594190.5 |
USHBP1
|
USH1 protein network component harmonin binding protein 1 |
| chr10_-_86957582 | 2.13 |
ENST00000372027.10
|
MMRN2
|
multimerin 2 |
| chr1_-_79188467 | 2.10 |
ENST00000656300.1
|
ADGRL4
|
adhesion G protein-coupled receptor L4 |
| chr6_+_54846735 | 2.09 |
ENST00000306858.8
|
FAM83B
|
family with sequence similarity 83 member B |
| chr1_-_12616762 | 2.07 |
ENST00000464917.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr5_+_42423433 | 2.04 |
ENST00000230882.9
|
GHR
|
growth hormone receptor |
| chr11_+_27040725 | 2.03 |
ENST00000529202.5
ENST00000263182.8 |
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr12_-_6124662 | 2.01 |
ENST00000261405.10
|
VWF
|
von Willebrand factor |
| chr6_+_36676455 | 2.01 |
ENST00000615513.4
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr17_-_41620801 | 2.01 |
ENST00000648859.1
|
KRT17
|
keratin 17 |
| chr7_+_80133830 | 1.99 |
ENST00000648098.1
ENST00000648476.1 ENST00000648412.1 ENST00000648953.1 ENST00000648306.1 ENST00000648832.1 ENST00000648877.1 ENST00000442586.2 ENST00000649487.1 ENST00000649267.1 |
GNAI1
|
G protein subunit alpha i1 |
| chr17_-_4560564 | 1.99 |
ENST00000574584.1
ENST00000381550.8 ENST00000301395.7 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr11_-_128867364 | 1.98 |
ENST00000440599.6
ENST00000324036.7 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr19_+_41219177 | 1.97 |
ENST00000301178.9
|
AXL
|
AXL receptor tyrosine kinase |
| chr16_+_66379225 | 1.97 |
ENST00000563425.2
|
CDH5
|
cadherin 5 |
| chr6_+_133889105 | 1.95 |
ENST00000367882.5
|
TCF21
|
transcription factor 21 |
| chr4_+_48986268 | 1.92 |
ENST00000226432.9
|
CWH43
|
cell wall biogenesis 43 C-terminal homolog |
| chr6_+_148342759 | 1.91 |
ENST00000367467.8
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr16_+_57619684 | 1.90 |
ENST00000568791.5
ENST00000570044.5 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr8_+_73294594 | 1.88 |
ENST00000240285.10
|
RDH10
|
retinol dehydrogenase 10 |
| chr12_-_57237090 | 1.86 |
ENST00000556732.1
|
NDUFA4L2
|
NDUFA4 mitochondrial complex associated like 2 |
| chr11_-_128867268 | 1.85 |
ENST00000392665.6
ENST00000392666.6 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr8_-_48921419 | 1.85 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2 |
| chr3_+_138347648 | 1.85 |
ENST00000614350.4
ENST00000289104.8 |
MRAS
|
muscle RAS oncogene homolog |
| chr15_+_40844171 | 1.83 |
ENST00000563656.5
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr19_-_46784905 | 1.83 |
ENST00000594991.5
|
SLC1A5
|
solute carrier family 1 member 5 |
| chrX_-_154354889 | 1.75 |
ENST00000438732.2
|
FLNA
|
filamin A |
| chr6_+_36676489 | 1.74 |
ENST00000448526.6
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr12_-_52493250 | 1.73 |
ENST00000330722.7
|
KRT6A
|
keratin 6A |
| chr16_+_57619709 | 1.66 |
ENST00000567553.5
ENST00000565314.5 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr19_-_5292770 | 1.66 |
ENST00000586065.1
|
PTPRS
|
protein tyrosine phosphatase receptor type S |
| chr20_-_6123019 | 1.65 |
ENST00000217289.9
ENST00000536936.1 |
FERMT1
|
fermitin family member 1 |
| chr20_+_36091409 | 1.62 |
ENST00000202028.9
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr1_-_182604379 | 1.60 |
ENST00000367558.6
|
RGS16
|
regulator of G protein signaling 16 |
| chr19_-_46413548 | 1.59 |
ENST00000307522.5
|
CCDC8
|
coiled-coil domain containing 8 |
| chr13_-_48413105 | 1.57 |
ENST00000620633.5
|
LPAR6
|
lysophosphatidic acid receptor 6 |
| chr1_+_17249088 | 1.57 |
ENST00000375460.3
|
PADI3
|
peptidyl arginine deiminase 3 |
| chrX_-_66639022 | 1.56 |
ENST00000374719.8
|
EDA2R
|
ectodysplasin A2 receptor |
| chr11_-_128842467 | 1.55 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr16_-_67966793 | 1.55 |
ENST00000541864.6
|
SLC12A4
|
solute carrier family 12 member 4 |
| chr19_-_17264732 | 1.50 |
ENST00000252597.8
|
USHBP1
|
USH1 protein network component harmonin binding protein 1 |
| chr1_-_93847150 | 1.49 |
ENST00000370244.5
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr18_+_63476927 | 1.47 |
ENST00000489441.5
ENST00000382771.9 ENST00000424602.1 |
SERPINB5
|
serpin family B member 5 |
| chr19_-_46784733 | 1.45 |
ENST00000593713.1
ENST00000598022.1 ENST00000434726.6 |
SLC1A5
|
solute carrier family 1 member 5 |
| chr1_-_79188390 | 1.43 |
ENST00000662530.1
|
ADGRL4
|
adhesion G protein-coupled receptor L4 |
| chr1_-_45623967 | 1.41 |
ENST00000445048.2
ENST00000421127.6 ENST00000528266.6 |
CCDC17
|
coiled-coil domain containing 17 |
| chr5_-_42811884 | 1.38 |
ENST00000514985.6
ENST00000511224.5 ENST00000507920.5 ENST00000510965.1 |
SELENOP
|
selenoprotein P |
| chr12_-_95790755 | 1.37 |
ENST00000343702.9
ENST00000344911.8 |
NTN4
|
netrin 4 |
| chr16_+_57619942 | 1.37 |
ENST00000568908.5
ENST00000568909.5 ENST00000566778.5 ENST00000561988.5 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr15_-_63156774 | 1.34 |
ENST00000462430.5
|
RPS27L
|
ribosomal protein S27 like |
| chr15_+_40844064 | 1.33 |
ENST00000568823.5
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr22_+_35648438 | 1.33 |
ENST00000409652.5
|
APOL6
|
apolipoprotein L6 |
| chr12_-_95791135 | 1.32 |
ENST00000538383.5
|
NTN4
|
netrin 4 |
| chr4_+_71339014 | 1.32 |
ENST00000340595.4
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr4_+_69096494 | 1.32 |
ENST00000508661.5
ENST00000622664.1 |
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr3_-_111595339 | 1.29 |
ENST00000317012.5
|
ZBED2
|
zinc finger BED-type containing 2 |
| chrX_+_67543973 | 1.27 |
ENST00000374690.9
|
AR
|
androgen receptor |
| chr1_+_116373233 | 1.27 |
ENST00000295598.10
|
ATP1A1
|
ATPase Na+/K+ transporting subunit alpha 1 |
| chr17_+_19648723 | 1.27 |
ENST00000672357.1
ENST00000584332.6 ENST00000176643.11 ENST00000339618.8 ENST00000579855.5 ENST00000671878.1 ENST00000395575.7 |
ALDH3A2
|
aldehyde dehydrogenase 3 family member A2 |
| chr12_-_52452139 | 1.25 |
ENST00000252252.4
|
KRT6B
|
keratin 6B |
| chr4_+_69096467 | 1.25 |
ENST00000305231.12
|
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr3_+_69866217 | 1.24 |
ENST00000314589.10
|
MITF
|
melanocyte inducing transcription factor |
| chr4_-_174522315 | 1.23 |
ENST00000514584.5
|
HPGD
|
15-hydroxyprostaglandin dehydrogenase |
| chr11_+_706595 | 1.21 |
ENST00000531348.5
ENST00000530636.5 |
EPS8L2
|
EPS8 like 2 |
| chr1_+_116372647 | 1.21 |
ENST00000418797.5
|
ATP1A1
|
ATPase Na+/K+ transporting subunit alpha 1 |
| chr12_+_119667859 | 1.20 |
ENST00000541640.5
|
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr4_-_10021490 | 1.20 |
ENST00000264784.8
|
SLC2A9
|
solute carrier family 2 member 9 |
| chr1_-_205850125 | 1.16 |
ENST00000367136.5
|
PM20D1
|
peptidase M20 domain containing 1 |
| chr11_+_111937320 | 1.16 |
ENST00000440460.7
|
DIXDC1
|
DIX domain containing 1 |
| chr12_+_119668109 | 1.16 |
ENST00000229328.10
ENST00000630317.1 |
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr4_-_128288163 | 1.14 |
ENST00000512483.5
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr17_+_19648915 | 1.14 |
ENST00000672567.1
ENST00000672709.1 |
ALDH3A2
|
aldehyde dehydrogenase 3 family member A2 |
| chr17_+_19648792 | 1.13 |
ENST00000630662.2
|
ALDH3A2
|
aldehyde dehydrogenase 3 family member A2 |
| chr8_-_23164020 | 1.13 |
ENST00000312584.4
|
TNFRSF10D
|
TNF receptor superfamily member 10d |
| chr1_+_26159071 | 1.11 |
ENST00000374268.5
|
FAM110D
|
family with sequence similarity 110 member D |
| chr8_-_23225061 | 1.11 |
ENST00000613472.1
ENST00000221132.8 |
TNFRSF10A
|
TNF receptor superfamily member 10a |
| chr3_-_185254028 | 1.11 |
ENST00000440662.1
ENST00000456310.5 ENST00000231887.8 |
EHHADH
|
enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase |
| chr7_+_75395629 | 1.09 |
ENST00000323819.7
ENST00000430211.5 |
TRIM73
|
tripartite motif containing 73 |
| chr19_+_12064720 | 1.09 |
ENST00000550826.1
ENST00000439326.8 |
ZNF844
|
zinc finger protein 844 |
| chr15_+_55408479 | 1.07 |
ENST00000569691.2
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr5_-_157059109 | 1.07 |
ENST00000523175.6
ENST00000522693.5 |
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr11_-_74467531 | 1.06 |
ENST00000310128.9
ENST00000531854.5 ENST00000526855.1 ENST00000529425.5 |
KCNE3
|
potassium voltage-gated channel subfamily E regulatory subunit 3 |
| chr3_+_136957948 | 1.05 |
ENST00000329582.9
|
IL20RB
|
interleukin 20 receptor subunit beta |
| chr3_+_53168687 | 1.04 |
ENST00000650940.1
ENST00000654719.1 |
PRKCD
|
protein kinase C delta |
| chr20_+_58689124 | 1.04 |
ENST00000525967.5
ENST00000525817.5 |
NPEPL1
|
aminopeptidase like 1 |
| chr22_+_37805218 | 1.03 |
ENST00000340857.4
|
H1-0
|
H1.0 linker histone |
| chr19_+_15049469 | 1.03 |
ENST00000427043.4
|
CASP14
|
caspase 14 |
| chr19_-_18940289 | 1.02 |
ENST00000542541.6
ENST00000433218.6 |
HOMER3
|
homer scaffold protein 3 |
| chr14_-_106579223 | 1.01 |
ENST00000390626.2
|
IGHV5-51
|
immunoglobulin heavy variable 5-51 |
| chr15_-_55408467 | 1.01 |
ENST00000310958.10
|
CCPG1
|
cell cycle progression 1 |
| chr1_+_236395394 | 1.00 |
ENST00000359362.6
|
EDARADD
|
EDAR associated death domain |
| chr21_-_26090035 | 0.99 |
ENST00000448850.5
|
APP
|
amyloid beta precursor protein |
| chr14_+_24310134 | 0.98 |
ENST00000533293.2
ENST00000543919.1 |
LTB4R2
|
leukotriene B4 receptor 2 |
| chr15_-_89690676 | 0.98 |
ENST00000561224.5
ENST00000300056.8 |
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr11_-_67629900 | 0.95 |
ENST00000301490.8
ENST00000376693.3 |
NUDT8
|
nudix hydrolase 8 |
| chr17_-_28335421 | 0.94 |
ENST00000578122.5
ENST00000579419.5 ENST00000585313.5 ENST00000578985.5 ENST00000577498.1 ENST00000585089.5 ENST00000357896.7 ENST00000395418.8 ENST00000588477.5 |
IFT20
|
intraflagellar transport 20 |
| chr1_+_86424154 | 0.93 |
ENST00000370565.5
|
CLCA2
|
chloride channel accessory 2 |
| chr3_+_122325237 | 0.93 |
ENST00000264474.4
ENST00000479204.1 |
CSTA
|
cystatin A |
| chr6_+_18387326 | 0.92 |
ENST00000259939.4
|
RNF144B
|
ring finger protein 144B |
| chr16_+_82056423 | 0.91 |
ENST00000568090.5
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr19_+_35358460 | 0.91 |
ENST00000327809.5
|
FFAR3
|
free fatty acid receptor 3 |
| chr11_+_64306227 | 0.91 |
ENST00000405666.5
ENST00000468670.2 |
ESRRA
|
estrogen related receptor alpha |
| chr4_-_128288791 | 0.90 |
ENST00000613358.4
ENST00000520121.6 |
PGRMC2
|
progesterone receptor membrane component 2 |
| chr11_-_78139258 | 0.88 |
ENST00000530910.6
ENST00000681417.1 ENST00000681225.1 ENST00000681765.1 ENST00000525870.6 ENST00000530608.6 ENST00000532306.6 ENST00000376156.7 ENST00000681699.1 ENST00000681221.1 ENST00000679444.1 ENST00000680829.1 ENST00000680761.1 ENST00000680256.1 ENST00000525783.6 ENST00000529139.6 ENST00000680580.1 ENST00000526849.6 ENST00000680399.1 ENST00000527099.2 ENST00000681489.1 ENST00000299626.10 ENST00000679497.1 ENST00000680101.1 ENST00000532440.6 ENST00000530454.6 ENST00000525761.3 ENST00000681575.1 ENST00000679559.1 ENST00000680398.1 ENST00000615266.5 ENST00000680643.1 |
ALG8
|
ALG8 alpha-1,3-glucosyltransferase |
| chr7_+_140696665 | 0.86 |
ENST00000476279.5
ENST00000461457.1 ENST00000465506.5 |
NDUFB2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr4_+_71339037 | 0.85 |
ENST00000512686.5
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr2_-_201642653 | 0.85 |
ENST00000621467.4
|
TMEM237
|
transmembrane protein 237 |
| chr4_-_143905529 | 0.85 |
ENST00000358615.9
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr15_-_63157464 | 0.84 |
ENST00000330964.10
ENST00000635699.1 ENST00000439025.1 |
RPS27L
|
ribosomal protein S27 like |
| chr7_+_140696696 | 0.84 |
ENST00000247866.9
ENST00000464566.5 |
NDUFB2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr19_-_11738882 | 0.84 |
ENST00000586121.1
ENST00000431998.1 ENST00000341191.11 ENST00000440527.1 |
ZNF823
|
zinc finger protein 823 |
| chr2_-_31138429 | 0.84 |
ENST00000349752.10
|
GALNT14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr1_-_185317234 | 0.82 |
ENST00000367498.8
|
IVNS1ABP
|
influenza virus NS1A binding protein |
| chr16_-_57764340 | 0.82 |
ENST00000565270.1
|
KIFC3
|
kinesin family member C3 |
| chr11_-_76210736 | 0.81 |
ENST00000529461.1
|
WNT11
|
Wnt family member 11 |
| chr17_+_79074822 | 0.80 |
ENST00000311595.14
ENST00000579016.6 |
ENGASE
|
endo-beta-N-acetylglucosaminidase |
| chr16_+_57620077 | 0.80 |
ENST00000567835.5
ENST00000569372.5 ENST00000563548.5 ENST00000562003.5 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr5_-_136193143 | 0.79 |
ENST00000607574.2
|
SMIM32
|
small integral membrane protein 32 |
| chr7_-_85187039 | 0.78 |
ENST00000284136.11
|
SEMA3D
|
semaphorin 3D |
| chr17_+_74987581 | 0.78 |
ENST00000337231.5
|
CDR2L
|
cerebellar degeneration related protein 2 like |
| chr15_+_51829644 | 0.78 |
ENST00000308580.12
|
TMOD3
|
tropomodulin 3 |
| chr11_+_47586678 | 0.77 |
ENST00000538490.3
|
FAM180B
|
family with sequence similarity 180 member B |
| chr22_+_22697789 | 0.77 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr8_-_23069012 | 0.77 |
ENST00000347739.3
ENST00000276431.9 |
TNFRSF10B
|
TNF receptor superfamily member 10b |
| chr5_+_163437569 | 0.76 |
ENST00000512163.5
ENST00000393929.5 ENST00000510097.5 ENST00000340828.7 ENST00000511490.4 ENST00000510664.5 |
CCNG1
|
cyclin G1 |
| chr5_-_180803830 | 0.76 |
ENST00000427865.2
ENST00000514283.1 |
MGAT1
|
alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase |
| chrX_+_152917830 | 0.75 |
ENST00000318529.12
|
ZNF185
|
zinc finger protein 185 with LIM domain |
| chr19_+_12064781 | 0.75 |
ENST00000441304.2
|
ZNF844
|
zinc finger protein 844 |
| chr5_+_126462339 | 0.74 |
ENST00000502348.5
|
GRAMD2B
|
GRAM domain containing 2B |
| chr13_+_23180960 | 0.74 |
ENST00000218867.4
|
SGCG
|
sarcoglycan gamma |
| chr4_-_40514543 | 0.73 |
ENST00000513473.5
|
RBM47
|
RNA binding motif protein 47 |
| chr9_-_127937800 | 0.72 |
ENST00000373110.4
ENST00000314392.13 |
DPM2
|
dolichyl-phosphate mannosyltransferase subunit 2, regulatory |
| chr8_-_73294425 | 0.70 |
ENST00000396465.5
|
RPL7
|
ribosomal protein L7 |
| chrX_-_66639255 | 0.69 |
ENST00000451436.6
|
EDA2R
|
ectodysplasin A2 receptor |
| chr6_+_150683593 | 0.69 |
ENST00000644968.1
|
PLEKHG1
|
pleckstrin homology and RhoGEF domain containing G1 |
| chr1_-_37692205 | 0.67 |
ENST00000477060.1
ENST00000491981.5 ENST00000488137.5 ENST00000619962.1 |
C1orf109
|
chromosome 1 open reading frame 109 |
| chr17_-_35753211 | 0.67 |
ENST00000604641.6
|
GAS2L2
|
growth arrest specific 2 like 2 |
| chr3_+_184363351 | 0.67 |
ENST00000443489.5
|
POLR2H
|
RNA polymerase II, I and III subunit H |
| chr11_+_27040826 | 0.66 |
ENST00000533566.5
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr17_+_28335653 | 0.66 |
ENST00000578158.5
|
TNFAIP1
|
TNF alpha induced protein 1 |
| chr5_+_116084992 | 0.66 |
ENST00000274458.9
ENST00000632434.1 |
COMMD10
|
COMM domain containing 10 |
| chr15_-_55408018 | 0.65 |
ENST00000569205.5
|
CCPG1
|
cell cycle progression 1 |
| chr17_+_28335718 | 0.64 |
ENST00000226225.7
|
TNFAIP1
|
TNF alpha induced protein 1 |
| chr15_+_51829625 | 0.64 |
ENST00000558455.1
|
TMOD3
|
tropomodulin 3 |
| chr8_-_97277890 | 0.63 |
ENST00000322128.5
|
TSPYL5
|
TSPY like 5 |
| chr2_-_31138503 | 0.62 |
ENST00000430167.1
|
GALNT14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr12_-_52191981 | 0.61 |
ENST00000313234.9
|
KRT80
|
keratin 80 |
| chr7_-_73328082 | 0.60 |
ENST00000333149.7
|
TRIM50
|
tripartite motif containing 50 |
| chr1_+_153775357 | 0.59 |
ENST00000624995.4
|
SLC27A3
|
solute carrier family 27 member 3 |
| chr12_-_57765591 | 0.59 |
ENST00000546567.5
|
CYP27B1
|
cytochrome P450 family 27 subfamily B member 1 |
| chr3_-_88059042 | 0.58 |
ENST00000309534.10
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr12_-_52192007 | 0.58 |
ENST00000394815.3
|
KRT80
|
keratin 80 |
| chr10_+_87863595 | 0.58 |
ENST00000371953.8
|
PTEN
|
phosphatase and tensin homolog |
| chr2_-_177392673 | 0.57 |
ENST00000447413.1
ENST00000397057.6 ENST00000456746.5 ENST00000464747.5 |
ENSG00000213963.6
NFE2L2
|
novel transcript nuclear factor, erythroid 2 like 2 |
| chr3_+_184363387 | 0.57 |
ENST00000452961.5
|
POLR2H
|
RNA polymerase II, I and III subunit H |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP63
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.9 | 3.5 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.8 | 3.3 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.8 | 4.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.8 | 10.2 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.8 | 2.3 | GO:0070662 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) mast cell proliferation(GO:0070662) |
| 0.7 | 2.8 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.7 | 2.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.6 | 3.2 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.6 | 3.8 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.6 | 1.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.6 | 3.6 | GO:0090131 | glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.5 | 2.5 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.4 | 3.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.4 | 1.7 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.4 | 1.3 | GO:1902905 | positive regulation of fibril organization(GO:1902905) |
| 0.4 | 3.7 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.4 | 1.6 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.4 | 0.8 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.4 | 7.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.4 | 2.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.4 | 2.3 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.4 | 1.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.4 | 1.4 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.4 | 1.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.3 | 3.9 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.3 | 1.6 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 1.2 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.3 | 2.0 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.3 | 3.0 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 | 1.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 1.0 | GO:2000754 | regulation of phospholipid scramblase activity(GO:1900161) positive regulation of phospholipid scramblase activity(GO:1900163) regulation of glucosylceramide catabolic process(GO:2000752) positive regulation of glucosylceramide catabolic process(GO:2000753) regulation of sphingomyelin catabolic process(GO:2000754) positive regulation of sphingomyelin catabolic process(GO:2000755) |
| 0.3 | 2.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.3 | 1.3 | GO:0060748 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.2 | 2.0 | GO:0044375 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.2 | 5.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 2.8 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 2.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 4.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 6.0 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.2 | 2.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 2.5 | GO:0031944 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of hormone biosynthetic process(GO:0032353) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.2 | 1.9 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.2 | 1.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 2.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 0.5 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.2 | 1.0 | GO:0051563 | microglia differentiation(GO:0014004) microglia development(GO:0014005) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.2 | 2.4 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 3.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 4.4 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.2 | 2.0 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.6 | GO:0090071 | rhythmic synaptic transmission(GO:0060024) negative regulation of ribosome biogenesis(GO:0090071) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 1.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 4.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.3 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 1.9 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 2.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.7 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 2.0 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 1.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 1.0 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 2.0 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 3.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.8 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 3.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.3 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) mesodermal cell migration(GO:0008078) cardiac cell fate determination(GO:0060913) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.1 | 0.5 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 3.7 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.1 | 2.0 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 2.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.5 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.1 | 1.2 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.1 | 0.6 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.1 | 1.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.2 | GO:0060733 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.1 | 0.2 | GO:0030221 | basophil differentiation(GO:0030221) astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 3.4 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.1 | 0.2 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.1 | 2.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 1.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.5 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 0.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.1 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.3 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 | 1.0 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.1 | 4.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.5 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 1.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 2.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 2.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.5 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 1.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.5 | GO:1901739 | regulation of myoblast fusion(GO:1901739) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.6 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 2.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 1.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.6 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.5 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.9 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.3 | GO:2001205 | TNFSF11-mediated signaling pathway(GO:0071847) negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 4.3 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 2.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 3.0 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.2 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 3.1 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.4 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 2.3 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.7 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.8 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.5 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 1.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.3 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.7 | 3.7 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.7 | 2.0 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.6 | 4.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.5 | 3.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 3.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 0.9 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 5.7 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.2 | 0.7 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 1.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 3.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 4.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 8.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.0 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 2.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 3.7 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.5 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 0.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 2.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 2.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.0 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 1.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 2.0 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 2.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 2.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 5.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 9.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 3.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 2.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 6.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.0 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 1.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 2.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 3.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.5 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 3.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.9 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 7.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 1.3 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0046577 | long-chain-alcohol oxidase activity(GO:0046577) |
| 1.0 | 6.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.8 | 10.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.8 | 3.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.7 | 3.7 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.7 | 2.7 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.5 | 2.6 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.5 | 2.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.5 | 2.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 5.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 4.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.4 | 6.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 1.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.4 | 3.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.3 | 1.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 2.8 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.3 | 2.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.3 | 1.0 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.3 | 3.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 1.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.2 | 0.7 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 0.7 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.2 | 0.9 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 0.9 | GO:0004583 | alpha-1,3-mannosyltransferase activity(GO:0000033) dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.2 | 1.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 1.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 0.6 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 0.2 | 3.0 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 0.9 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 2.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 2.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 2.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 2.5 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 1.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.5 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 2.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 1.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 5.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 2.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.8 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 3.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 4.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.3 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 1.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 4.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.5 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 2.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 2.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 3.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 2.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 2.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 4.4 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.1 | 2.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.0 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 1.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.8 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 3.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 2.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 1.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 3.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.0 | 0.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 1.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.5 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 3.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 7.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 1.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 5.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.4 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 3.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.8 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.8 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 2.6 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 2.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 1.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 10.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 11.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 6.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 6.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 3.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 14.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 3.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 3.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.2 | 5.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 4.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 2.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 5.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 9.4 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 1.0 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 3.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 1.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 3.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 5.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 2.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |