Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
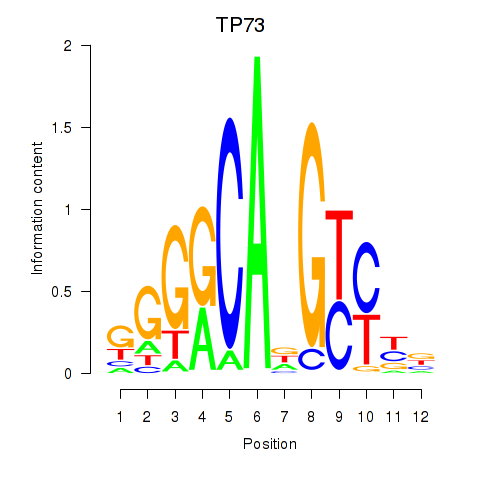
Results for TP73
Z-value: 0.81
Transcription factors associated with TP73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP73
|
ENSG00000078900.15 | tumor protein p73 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP73 | hg38_v1_chr1_+_3698027_3698058 | 0.24 | 1.9e-01 | Click! |
Activity profile of TP73 motif
Sorted Z-values of TP73 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_74174403 | 1.78 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine rich repeat |
| chr12_+_49295138 | 1.71 |
ENST00000257860.9
|
PRPH
|
peripherin |
| chr1_+_46671821 | 1.66 |
ENST00000334122.5
ENST00000415500.1 |
TEX38
|
testis expressed 38 |
| chr15_+_40844018 | 1.63 |
ENST00000344051.8
ENST00000562057.6 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr2_-_165794190 | 1.61 |
ENST00000392701.8
ENST00000422973.1 |
GALNT3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr2_+_176116768 | 1.49 |
ENST00000249501.5
|
HOXD10
|
homeobox D10 |
| chr1_+_46671871 | 1.40 |
ENST00000564373.1
|
TEX38
|
testis expressed 38 |
| chr2_+_219572304 | 1.38 |
ENST00000243786.3
|
INHA
|
inhibin subunit alpha |
| chr9_+_36169382 | 1.31 |
ENST00000335119.4
|
CCIN
|
calicin |
| chr19_+_44777860 | 1.30 |
ENST00000341505.4
ENST00000647358.2 |
CBLC
|
Cbl proto-oncogene C |
| chr2_+_95274439 | 1.27 |
ENST00000317620.14
ENST00000403131.6 ENST00000317668.8 |
PROM2
|
prominin 2 |
| chr15_+_40844506 | 1.11 |
ENST00000568580.5
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr1_-_153615858 | 1.10 |
ENST00000476873.5
|
S100A14
|
S100 calcium binding protein A14 |
| chr1_-_204190324 | 1.06 |
ENST00000638118.1
|
REN
|
renin |
| chr15_+_75206398 | 1.06 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr15_+_75206014 | 1.06 |
ENST00000567617.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr1_+_239386942 | 0.93 |
ENST00000675184.1
|
CHRM3
|
cholinergic receptor muscarinic 3 |
| chr19_+_43716070 | 0.92 |
ENST00000244314.6
|
IRGC
|
immunity related GTPase cinema |
| chr11_+_46277648 | 0.90 |
ENST00000621158.5
|
CREB3L1
|
cAMP responsive element binding protein 3 like 1 |
| chr10_-_103452356 | 0.88 |
ENST00000260743.10
|
CALHM2
|
calcium homeostasis modulator family member 2 |
| chr22_-_37580075 | 0.87 |
ENST00000215886.6
|
LGALS2
|
galectin 2 |
| chr15_+_40844171 | 0.86 |
ENST00000563656.5
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr19_+_3721719 | 0.86 |
ENST00000589378.5
|
TJP3
|
tight junction protein 3 |
| chr10_-_103452384 | 0.85 |
ENST00000369788.7
|
CALHM2
|
calcium homeostasis modulator family member 2 |
| chr11_+_60429567 | 0.85 |
ENST00000300190.7
|
MS4A5
|
membrane spanning 4-domains A5 |
| chr19_-_35490456 | 0.85 |
ENST00000338897.4
ENST00000484218.6 |
KRTDAP
|
keratinocyte differentiation associated protein |
| chr6_+_36676455 | 0.81 |
ENST00000615513.4
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chrX_+_9912434 | 0.81 |
ENST00000418909.6
|
SHROOM2
|
shroom family member 2 |
| chr6_+_36676489 | 0.78 |
ENST00000448526.6
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr2_+_219418369 | 0.78 |
ENST00000373960.4
|
DES
|
desmin |
| chr5_-_157166262 | 0.78 |
ENST00000302938.4
|
FAM71B
|
family with sequence similarity 71 member B |
| chr7_+_142939343 | 0.76 |
ENST00000458732.1
ENST00000409607.5 |
LLCFC1
|
LLLL and CFNLAS motif containing 1 |
| chr8_+_37796850 | 0.76 |
ENST00000412232.3
|
ADGRA2
|
adhesion G protein-coupled receptor A2 |
| chr8_+_119873710 | 0.74 |
ENST00000523492.5
ENST00000286234.6 |
DEPTOR
|
DEP domain containing MTOR interacting protein |
| chr19_+_43716095 | 0.74 |
ENST00000596627.1
|
IRGC
|
immunity related GTPase cinema |
| chr8_+_37796906 | 0.73 |
ENST00000315215.11
|
ADGRA2
|
adhesion G protein-coupled receptor A2 |
| chr10_-_77926724 | 0.73 |
ENST00000372391.7
|
DLG5
|
discs large MAGUK scaffold protein 5 |
| chr2_-_229271221 | 0.72 |
ENST00000392054.7
ENST00000409462.1 ENST00000392055.8 |
PID1
|
phosphotyrosine interaction domain containing 1 |
| chr2_-_26641363 | 0.72 |
ENST00000288861.5
|
CIB4
|
calcium and integrin binding family member 4 |
| chr9_+_35673917 | 0.72 |
ENST00000617161.1
ENST00000378357.9 |
CA9
|
carbonic anhydrase 9 |
| chrX_-_81121663 | 0.71 |
ENST00000430960.5
ENST00000447319.5 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr11_+_77589911 | 0.69 |
ENST00000313578.4
|
AQP11
|
aquaporin 11 |
| chr12_-_91179472 | 0.68 |
ENST00000550099.5
ENST00000546391.5 |
DCN
|
decorin |
| chr5_+_38258373 | 0.67 |
ENST00000354891.7
|
EGFLAM
|
EGF like, fibronectin type III and laminin G domains |
| chr19_+_35106510 | 0.66 |
ENST00000648240.1
|
ENSG00000285526.1
|
novel protein |
| chr19_+_44751251 | 0.66 |
ENST00000444487.1
|
BCL3
|
BCL3 transcription coactivator |
| chr5_+_38258551 | 0.65 |
ENST00000322350.10
|
EGFLAM
|
EGF like, fibronectin type III and laminin G domains |
| chr4_+_38664189 | 0.64 |
ENST00000514033.1
ENST00000261438.10 |
KLF3
|
Kruppel like factor 3 |
| chr1_-_236065079 | 0.62 |
ENST00000264187.7
ENST00000366595.7 |
NID1
|
nidogen 1 |
| chr17_+_74431338 | 0.61 |
ENST00000342648.9
ENST00000652232.1 ENST00000481232.2 |
GPRC5C
|
G protein-coupled receptor class C group 5 member C |
| chr10_+_68901258 | 0.61 |
ENST00000373585.8
|
DDX50
|
DExD-box helicase 50 |
| chr1_-_68232514 | 0.60 |
ENST00000262348.9
ENST00000370973.2 ENST00000370971.1 |
WLS
|
Wnt ligand secretion mediator |
| chr1_-_68232539 | 0.60 |
ENST00000370976.7
ENST00000354777.6 |
WLS
|
Wnt ligand secretion mediator |
| chr16_+_84145256 | 0.58 |
ENST00000378553.10
|
DNAAF1
|
dynein axonemal assembly factor 1 |
| chr9_-_135499846 | 0.58 |
ENST00000429260.7
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chrX_-_81121619 | 0.58 |
ENST00000373250.7
|
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chrX_+_153642473 | 0.56 |
ENST00000370167.8
|
DUSP9
|
dual specificity phosphatase 9 |
| chr11_+_31812307 | 0.55 |
ENST00000643436.1
ENST00000646959.1 ENST00000645942.1 ENST00000530348.5 |
PAUPAR
ENSG00000285283.1
|
PAX6 upstream antisense RNA novel protein |
| chr2_+_69741974 | 0.55 |
ENST00000409920.5
|
ANXA4
|
annexin A4 |
| chr1_+_27959943 | 0.53 |
ENST00000675575.1
ENST00000373884.6 |
XKR8
|
XK related 8 |
| chrX_+_131058340 | 0.53 |
ENST00000276211.10
ENST00000370922.5 |
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr1_+_239386556 | 0.52 |
ENST00000676153.1
|
CHRM3
|
cholinergic receptor muscarinic 3 |
| chr3_+_136957948 | 0.52 |
ENST00000329582.9
|
IL20RB
|
interleukin 20 receptor subunit beta |
| chr17_-_76027296 | 0.52 |
ENST00000301607.8
|
EVPL
|
envoplakin |
| chr1_+_100352925 | 0.51 |
ENST00000644813.1
|
CDC14A
|
cell division cycle 14A |
| chr14_+_52314280 | 0.50 |
ENST00000557436.1
ENST00000245457.6 |
PTGER2
|
prostaglandin E receptor 2 |
| chr7_+_69967464 | 0.49 |
ENST00000664521.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr17_+_39200507 | 0.49 |
ENST00000678573.1
|
RPL19
|
ribosomal protein L19 |
| chr6_+_36130586 | 0.49 |
ENST00000373759.1
|
MAPK13
|
mitogen-activated protein kinase 13 |
| chr19_+_41860236 | 0.48 |
ENST00000601492.5
ENST00000600467.6 ENST00000598742.6 ENST00000221975.6 ENST00000598261.2 |
RPS19
|
ribosomal protein S19 |
| chr4_-_156970903 | 0.48 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr17_+_32928126 | 0.48 |
ENST00000579849.6
ENST00000578289.5 ENST00000439138.5 |
TMEM98
|
transmembrane protein 98 |
| chrX_+_9912548 | 0.48 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr17_+_39200275 | 0.47 |
ENST00000225430.9
|
RPL19
|
ribosomal protein L19 |
| chr17_+_39200334 | 0.47 |
ENST00000579260.5
ENST00000582193.5 |
RPL19
|
ribosomal protein L19 |
| chr17_+_54900824 | 0.46 |
ENST00000572405.5
ENST00000572158.5 ENST00000575882.6 ENST00000572298.5 ENST00000536554.5 ENST00000575333.5 ENST00000570499.5 ENST00000572576.5 |
TOM1L1
|
target of myb1 like 1 membrane trafficking protein |
| chr17_-_44123628 | 0.46 |
ENST00000587135.1
ENST00000225983.10 ENST00000682912.1 ENST00000588703.5 |
HDAC5
|
histone deacetylase 5 |
| chr15_+_40844064 | 0.45 |
ENST00000568823.5
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr17_+_7888783 | 0.43 |
ENST00000330494.12
ENST00000358181.8 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr9_-_114505437 | 0.43 |
ENST00000374057.3
ENST00000362057.4 ENST00000673697.1 |
WHRN
|
whirlin |
| chr17_-_76027212 | 0.43 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr1_-_203086001 | 0.42 |
ENST00000241651.5
|
MYOG
|
myogenin |
| chr16_+_89918855 | 0.41 |
ENST00000555147.2
|
MC1R
|
melanocortin 1 receptor |
| chr17_-_5123102 | 0.40 |
ENST00000250076.7
|
ZNF232
|
zinc finger protein 232 |
| chr17_+_39200302 | 0.39 |
ENST00000579374.5
|
RPL19
|
ribosomal protein L19 |
| chr9_-_20382461 | 0.38 |
ENST00000380321.5
ENST00000629733.3 |
MLLT3
|
MLLT3 super elongation complex subunit |
| chr14_-_74612021 | 0.38 |
ENST00000556690.5
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr3_-_185821092 | 0.38 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr17_-_81514629 | 0.37 |
ENST00000681052.1
ENST00000575659.6 |
ACTG1
|
actin gamma 1 |
| chr10_+_46375619 | 0.37 |
ENST00000584982.7
ENST00000613703.4 |
ANXA8L1
|
annexin A8 like 1 |
| chr17_+_56153458 | 0.35 |
ENST00000318698.6
ENST00000682825.1 ENST00000566473.6 |
ANKFN1
|
ankyrin repeat and fibronectin type III domain containing 1 |
| chr4_+_109560219 | 0.34 |
ENST00000394650.7
|
MCUB
|
mitochondrial calcium uniporter dominant negative subunit beta |
| chr3_-_171026709 | 0.33 |
ENST00000314251.8
|
SLC2A2
|
solute carrier family 2 member 2 |
| chr3_+_44976236 | 0.32 |
ENST00000265564.8
|
EXOSC7
|
exosome component 7 |
| chr9_-_129620743 | 0.31 |
ENST00000619117.1
ENST00000372478.4 |
C9orf50
|
chromosome 9 open reading frame 50 |
| chr14_+_21749163 | 0.31 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr7_-_128405930 | 0.30 |
ENST00000470772.5
ENST00000480861.5 ENST00000496200.5 |
IMPDH1
|
inosine monophosphate dehydrogenase 1 |
| chr12_+_82358496 | 0.29 |
ENST00000248306.8
ENST00000548200.5 |
METTL25
|
methyltransferase like 25 |
| chr12_-_82358380 | 0.28 |
ENST00000256151.8
ENST00000552377.5 |
CCDC59
|
coiled-coil domain containing 59 |
| chr6_-_33288887 | 0.28 |
ENST00000444176.1
|
WDR46
|
WD repeat domain 46 |
| chr19_+_41350911 | 0.28 |
ENST00000539627.5
|
TMEM91
|
transmembrane protein 91 |
| chr11_+_60429595 | 0.27 |
ENST00000528905.1
ENST00000528093.1 |
MS4A5
|
membrane spanning 4-domains A5 |
| chr2_-_38602626 | 0.26 |
ENST00000378915.7
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L like |
| chr14_-_89954659 | 0.25 |
ENST00000555070.1
ENST00000316738.12 ENST00000538485.6 ENST00000556609.5 |
ENSG00000259053.1
EFCAB11
|
novel transcript EF-hand calcium binding domain 11 |
| chr16_+_2019777 | 0.25 |
ENST00000566435.4
|
NPW
|
neuropeptide W |
| chr2_+_127535746 | 0.24 |
ENST00000428314.5
ENST00000409816.7 |
MYO7B
|
myosin VIIB |
| chr17_-_40867200 | 0.24 |
ENST00000647902.1
ENST00000251643.5 |
KRT12
|
keratin 12 |
| chr9_+_6757940 | 0.24 |
ENST00000381309.8
|
KDM4C
|
lysine demethylase 4C |
| chr10_-_60140515 | 0.23 |
ENST00000486349.2
|
ANK3
|
ankyrin 3 |
| chr20_-_54173516 | 0.23 |
ENST00000395955.7
|
CYP24A1
|
cytochrome P450 family 24 subfamily A member 1 |
| chr17_+_67825664 | 0.23 |
ENST00000321892.8
|
BPTF
|
bromodomain PHD finger transcription factor |
| chr1_+_16043776 | 0.22 |
ENST00000375679.9
|
CLCNKB
|
chloride voltage-gated channel Kb |
| chr10_+_46375721 | 0.22 |
ENST00000616785.1
ENST00000611655.1 ENST00000619162.5 |
ENSG00000285402.1
ANXA8L1
|
novel transcript annexin A8 like 1 |
| chr4_+_105710809 | 0.22 |
ENST00000360505.9
ENST00000510865.5 ENST00000509336.5 |
GSTCD
|
glutathione S-transferase C-terminal domain containing |
| chr6_+_43576205 | 0.21 |
ENST00000372226.1
ENST00000443535.1 |
POLH
|
DNA polymerase eta |
| chr15_+_64387828 | 0.21 |
ENST00000261884.8
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr12_+_43758936 | 0.20 |
ENST00000440781.6
ENST00000431837.5 ENST00000550616.5 ENST00000613694.5 ENST00000551736.5 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr1_-_160862880 | 0.20 |
ENST00000368034.9
|
CD244
|
CD244 molecule |
| chr6_-_33289189 | 0.20 |
ENST00000374617.9
|
WDR46
|
WD repeat domain 46 |
| chr17_-_49414802 | 0.20 |
ENST00000511832.6
ENST00000419140.7 ENST00000617874.5 |
PHB
|
prohibitin |
| chr20_+_41136944 | 0.19 |
ENST00000244007.7
|
PLCG1
|
phospholipase C gamma 1 |
| chr19_-_13937991 | 0.19 |
ENST00000254320.7
ENST00000586075.1 |
PODNL1
|
podocan like 1 |
| chr1_-_235161215 | 0.19 |
ENST00000447801.5
ENST00000408888.8 ENST00000429912.1 |
RBM34
|
RNA binding motif protein 34 |
| chr19_-_35742431 | 0.18 |
ENST00000592537.5
ENST00000246532.6 ENST00000588992.5 |
IGFLR1
|
IGF like family receptor 1 |
| chr16_-_14630200 | 0.18 |
ENST00000650990.1
ENST00000651049.1 ENST00000652727.1 ENST00000652501.1 ENST00000539279.5 ENST00000651634.1 ENST00000651027.1 ENST00000420015.6 ENST00000651865.1 ENST00000437198.7 ENST00000341484.11 ENST00000650960.1 |
PARN
|
poly(A)-specific ribonuclease |
| chr11_+_71527267 | 0.17 |
ENST00000398536.6
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr10_+_46375645 | 0.16 |
ENST00000622769.4
|
ANXA8L1
|
annexin A8 like 1 |
| chr10_-_89535575 | 0.15 |
ENST00000371790.5
|
SLC16A12
|
solute carrier family 16 member 12 |
| chr1_+_16043736 | 0.15 |
ENST00000619181.4
|
CLCNKB
|
chloride voltage-gated channel Kb |
| chr14_+_75278820 | 0.15 |
ENST00000554617.1
ENST00000554212.5 ENST00000535987.5 ENST00000303562.9 ENST00000555242.1 |
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr16_+_30923565 | 0.14 |
ENST00000338343.10
|
FBXL19
|
F-box and leucine rich repeat protein 19 |
| chr4_+_6716131 | 0.14 |
ENST00000320776.5
|
BLOC1S4
|
biogenesis of lysosomal organelles complex 1 subunit 4 |
| chr13_+_50910018 | 0.14 |
ENST00000645990.1
ENST00000642995.1 ENST00000636524.2 ENST00000643159.1 ENST00000645618.1 ENST00000611510.5 ENST00000642454.1 ENST00000646709.1 |
RNASEH2B
|
ribonuclease H2 subunit B |
| chr17_-_49414871 | 0.14 |
ENST00000504124.6
ENST00000300408.8 ENST00000614445.5 |
PHB
|
prohibitin |
| chr6_+_31547560 | 0.14 |
ENST00000376148.9
ENST00000376145.8 |
NFKBIL1
|
NFKB inhibitor like 1 |
| chr17_+_67825494 | 0.13 |
ENST00000306378.11
ENST00000544778.6 |
BPTF
|
bromodomain PHD finger transcription factor |
| chr16_-_67966793 | 0.13 |
ENST00000541864.6
|
SLC12A4
|
solute carrier family 12 member 4 |
| chr1_-_51878711 | 0.13 |
ENST00000352171.12
|
NRDC
|
nardilysin convertase |
| chr1_-_160862700 | 0.12 |
ENST00000322302.7
ENST00000368033.7 |
CD244
|
CD244 molecule |
| chr14_+_22086401 | 0.11 |
ENST00000390451.2
|
TRAV23DV6
|
T cell receptor alpha variable 23/delta variable 6 |
| chr19_-_38735405 | 0.11 |
ENST00000597987.5
ENST00000595177.1 |
CAPN12
|
calpain 12 |
| chr9_-_20382448 | 0.10 |
ENST00000491137.5
|
MLLT3
|
MLLT3 super elongation complex subunit |
| chr9_+_128683645 | 0.10 |
ENST00000372692.8
|
SET
|
SET nuclear proto-oncogene |
| chr17_-_30334050 | 0.09 |
ENST00000328886.5
ENST00000538566.6 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr19_+_43533384 | 0.09 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr8_-_144063951 | 0.08 |
ENST00000567871.2
|
OPLAH
|
5-oxoprolinase, ATP-hydrolysing |
| chr14_+_75279637 | 0.08 |
ENST00000555686.1
ENST00000555672.1 |
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chrX_+_154542194 | 0.07 |
ENST00000618670.4
|
IKBKG
|
inhibitor of nuclear factor kappa B kinase regulatory subunit gamma |
| chr20_-_54173976 | 0.07 |
ENST00000216862.8
|
CYP24A1
|
cytochrome P450 family 24 subfamily A member 1 |
| chr10_+_62804710 | 0.07 |
ENST00000373783.3
|
ADO
|
2-aminoethanethiol dioxygenase |
| chr10_-_97426044 | 0.07 |
ENST00000536831.5
ENST00000622320.4 ENST00000439965.6 |
RRP12
|
ribosomal RNA processing 12 homolog |
| chr1_-_182672232 | 0.05 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr22_+_35383106 | 0.05 |
ENST00000678411.1
|
HMOX1
|
heme oxygenase 1 |
| chr6_+_43576119 | 0.05 |
ENST00000372236.9
|
POLH
|
DNA polymerase eta |
| chr1_-_220046432 | 0.04 |
ENST00000609181.5
ENST00000366923.8 |
EPRS1
|
glutamyl-prolyl-tRNA synthetase 1 |
| chr19_-_1863497 | 0.04 |
ENST00000617223.1
ENST00000250916.6 |
KLF16
|
Kruppel like factor 16 |
| chrX_-_132094263 | 0.04 |
ENST00000370879.5
|
FRMD7
|
FERM domain containing 7 |
| chrX_+_73447042 | 0.03 |
ENST00000373514.3
|
CDX4
|
caudal type homeobox 4 |
| chr2_-_27134611 | 0.02 |
ENST00000406567.7
ENST00000260643.7 |
PREB
|
prolactin regulatory element binding |
| chrX_-_139642518 | 0.02 |
ENST00000370573.8
ENST00000338585.6 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr1_+_156815633 | 0.02 |
ENST00000392302.7
ENST00000674537.1 |
NTRK1
|
neurotrophic receptor tyrosine kinase 1 |
| chr17_+_21288029 | 0.02 |
ENST00000526076.6
ENST00000361818.9 ENST00000316920.10 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr11_+_111977300 | 0.02 |
ENST00000615255.1
|
DIXDC1
|
DIX domain containing 1 |
| chr1_-_77979054 | 0.02 |
ENST00000370768.7
ENST00000370767.5 ENST00000421641.1 |
FUBP1
|
far upstream element binding protein 1 |
| chr9_+_113580912 | 0.02 |
ENST00000471324.6
|
RGS3
|
regulator of G protein signaling 3 |
| chr1_-_51878799 | 0.02 |
ENST00000354831.11
ENST00000544028.5 |
NRDC
|
nardilysin convertase |
| chr19_+_12995554 | 0.01 |
ENST00000397661.6
|
NFIX
|
nuclear factor I X |
| chr9_+_6758133 | 0.01 |
ENST00000543771.5
|
KDM4C
|
lysine demethylase 4C |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP73
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.4 | 1.3 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.3 | 1.4 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.3 | 1.6 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.3 | 1.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.7 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.2 | 1.5 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.2 | 0.7 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 1.6 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.2 | 1.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.2 | 0.5 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.2 | 1.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.2 | 0.5 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.4 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 4.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.9 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.3 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.3 | GO:0071663 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.1 | 0.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 2.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.3 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.3 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.5 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 1.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.2 | GO:1902598 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.5 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.3 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.5 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 1.9 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 1.0 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.7 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.8 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.8 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.5 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.5 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.7 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 1.3 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.3 | 1.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 1.3 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 1.5 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 0.7 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.2 | GO:0090651 | apical cytoplasm(GO:0090651) |
| 0.1 | 0.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 1.6 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 1.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 1.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.2 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 4.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.1 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 2.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |