Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for UAUUGCU
Z-value: 1.48
miRNA associated with seed UAUUGCU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-137
|
MIMAT0000429 |
Activity profile of UAUUGCU motif
Sorted Z-values of UAUUGCU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_236686454 | 7.64 |
ENST00000542672.6
ENST00000366578.6 ENST00000682015.1 ENST00000651275.1 |
ACTN2
|
actinin alpha 2 |
| chr5_-_150289764 | 5.38 |
ENST00000671881.1
ENST00000672752.1 ENST00000510347.2 ENST00000672829.1 ENST00000348628.11 |
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr19_-_2721332 | 4.54 |
ENST00000588128.1
ENST00000323469.5 |
DIRAS1
|
DIRAS family GTPase 1 |
| chr1_-_32872473 | 4.13 |
ENST00000496770.1
|
FNDC5
|
fibronectin type III domain containing 5 |
| chr5_+_7396099 | 4.11 |
ENST00000338316.9
|
ADCY2
|
adenylate cyclase 2 |
| chr8_-_123541197 | 4.03 |
ENST00000517956.5
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr13_+_23180960 | 3.97 |
ENST00000218867.4
|
SGCG
|
sarcoglycan gamma |
| chr9_-_98708856 | 3.84 |
ENST00000259455.4
|
GABBR2
|
gamma-aminobutyric acid type B receptor subunit 2 |
| chr1_+_237042176 | 3.64 |
ENST00000366574.7
|
RYR2
|
ryanodine receptor 2 |
| chr3_-_62875337 | 3.58 |
ENST00000357948.7
ENST00000612439.4 ENST00000383710.9 |
CADPS
|
calcium dependent secretion activator |
| chr13_-_25172278 | 3.32 |
ENST00000515384.2
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr6_+_17281341 | 3.28 |
ENST00000379052.10
|
RBM24
|
RNA binding motif protein 24 |
| chr17_-_76240478 | 3.27 |
ENST00000269391.11
|
RNF157
|
ring finger protein 157 |
| chr9_-_90642791 | 3.23 |
ENST00000375765.5
ENST00000636786.1 |
DIRAS2
|
DIRAS family GTPase 2 |
| chr19_-_11481044 | 3.22 |
ENST00000359227.8
|
ELAVL3
|
ELAV like RNA binding protein 3 |
| chr8_-_22156789 | 3.18 |
ENST00000306317.7
|
LGI3
|
leucine rich repeat LGI family member 3 |
| chr11_+_109422174 | 3.07 |
ENST00000327419.7
|
C11orf87
|
chromosome 11 open reading frame 87 |
| chr11_-_47352693 | 2.98 |
ENST00000256993.8
ENST00000399249.6 ENST00000545968.6 |
MYBPC3
|
myosin binding protein C3 |
| chr2_+_206443496 | 2.97 |
ENST00000264377.8
|
ADAM23
|
ADAM metallopeptidase domain 23 |
| chr3_+_10992717 | 2.97 |
ENST00000642767.1
ENST00000425938.6 ENST00000642515.1 ENST00000643498.1 ENST00000646072.1 ENST00000646570.1 ENST00000287766.10 ENST00000645281.1 ENST00000642820.1 ENST00000645054.1 ENST00000642735.1 ENST00000646022.1 ENST00000645776.1 ENST00000645592.1 ENST00000646924.1 ENST00000645974.1 ENST00000644314.1 ENST00000642639.1 ENST00000646060.1 ENST00000642201.1 ENST00000646487.1 ENST00000647194.1 ENST00000646088.1 |
SLC6A1
|
solute carrier family 6 member 1 |
| chr14_+_92923143 | 2.86 |
ENST00000216492.10
ENST00000334654.4 |
CHGA
|
chromogranin A |
| chr15_+_84817346 | 2.84 |
ENST00000258888.6
|
ALPK3
|
alpha kinase 3 |
| chr1_+_95117324 | 2.73 |
ENST00000370203.9
ENST00000456991.5 |
TLCD4
|
TLC domain containing 4 |
| chr4_-_52659238 | 2.73 |
ENST00000451218.6
ENST00000441222.8 |
USP46
|
ubiquitin specific peptidase 46 |
| chr6_-_89412219 | 2.69 |
ENST00000369415.9
|
RRAGD
|
Ras related GTP binding D |
| chr12_+_4909895 | 2.66 |
ENST00000638821.1
ENST00000382545.5 |
ENSG00000256654.4
KCNA1
|
novel transcript, sense overlapping KCNA1 potassium voltage-gated channel subfamily A member 1 |
| chr4_-_140427635 | 2.61 |
ENST00000325617.10
ENST00000414773.5 |
CLGN
|
calmegin |
| chr1_+_110150480 | 2.58 |
ENST00000331565.5
|
SLC6A17
|
solute carrier family 6 member 17 |
| chr10_+_25174969 | 2.51 |
ENST00000376351.4
|
GPR158
|
G protein-coupled receptor 158 |
| chr9_-_34522968 | 2.49 |
ENST00000399775.3
|
ENHO
|
energy homeostasis associated |
| chr1_-_193186599 | 2.45 |
ENST00000367434.5
|
B3GALT2
|
beta-1,3-galactosyltransferase 2 |
| chr11_-_61581104 | 2.42 |
ENST00000263846.8
|
SYT7
|
synaptotagmin 7 |
| chr4_-_152536045 | 2.37 |
ENST00000603548.6
ENST00000281708.10 |
FBXW7
|
F-box and WD repeat domain containing 7 |
| chr22_+_41976933 | 2.34 |
ENST00000396425.7
|
SEPTIN3
|
septin 3 |
| chr3_+_49554436 | 2.33 |
ENST00000296452.5
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr20_-_33674359 | 2.29 |
ENST00000606690.5
ENST00000439478.5 ENST00000246190.11 ENST00000375238.8 |
NECAB3
|
N-terminal EF-hand calcium binding protein 3 |
| chr14_-_34713788 | 2.28 |
ENST00000341223.8
|
CFL2
|
cofilin 2 |
| chr20_-_49482645 | 2.26 |
ENST00000371741.6
|
KCNB1
|
potassium voltage-gated channel subfamily B member 1 |
| chr11_+_22338333 | 2.25 |
ENST00000263160.4
|
SLC17A6
|
solute carrier family 17 member 6 |
| chr9_-_89178810 | 2.22 |
ENST00000375835.9
|
SHC3
|
SHC adaptor protein 3 |
| chr20_+_24469623 | 2.20 |
ENST00000376862.4
|
SYNDIG1
|
synapse differentiation inducing 1 |
| chr6_-_46325641 | 2.18 |
ENST00000330430.10
ENST00000405162.2 |
RCAN2
|
regulator of calcineurin 2 |
| chr18_-_72867945 | 2.15 |
ENST00000327305.11
|
NETO1
|
neuropilin and tolloid like 1 |
| chr12_+_51591216 | 2.15 |
ENST00000668547.1
ENST00000354534.11 ENST00000627620.5 ENST00000545061.5 |
SCN8A
|
sodium voltage-gated channel alpha subunit 8 |
| chr15_+_90001300 | 2.14 |
ENST00000268154.9
|
ZNF710
|
zinc finger protein 710 |
| chr5_+_31639104 | 2.14 |
ENST00000438447.2
|
PDZD2
|
PDZ domain containing 2 |
| chr9_-_19786928 | 2.13 |
ENST00000341998.6
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 member 2 |
| chr14_+_64704380 | 2.09 |
ENST00000247226.13
ENST00000394691.7 |
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr1_-_118989504 | 2.07 |
ENST00000207157.7
|
TBX15
|
T-box transcription factor 15 |
| chr19_+_589873 | 2.05 |
ENST00000251287.3
|
HCN2
|
hyperpolarization activated cyclic nucleotide gated potassium and sodium channel 2 |
| chr22_+_29480211 | 2.04 |
ENST00000310624.7
|
NEFH
|
neurofilament heavy |
| chr2_-_50347710 | 2.03 |
ENST00000342183.9
ENST00000401710.5 |
NRXN1
|
neurexin 1 |
| chr6_-_166382927 | 1.99 |
ENST00000360961.11
|
MPC1
|
mitochondrial pyruvate carrier 1 |
| chr3_-_9249623 | 1.99 |
ENST00000383836.8
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr20_-_62407274 | 1.97 |
ENST00000279101.9
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2 |
| chrX_+_153687918 | 1.95 |
ENST00000253122.10
|
SLC6A8
|
solute carrier family 6 member 8 |
| chr11_-_790062 | 1.95 |
ENST00000330106.5
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr15_-_65517244 | 1.95 |
ENST00000341861.9
|
DPP8
|
dipeptidyl peptidase 8 |
| chr11_+_24497155 | 1.95 |
ENST00000529015.5
ENST00000533227.5 |
LUZP2
|
leucine zipper protein 2 |
| chr12_-_75209422 | 1.95 |
ENST00000393288.2
ENST00000540018.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr3_+_84958963 | 1.93 |
ENST00000383699.8
|
CADM2
|
cell adhesion molecule 2 |
| chr8_-_40897814 | 1.91 |
ENST00000297737.11
ENST00000315769.11 |
ZMAT4
|
zinc finger matrin-type 4 |
| chr5_+_80035341 | 1.90 |
ENST00000350881.6
|
THBS4
|
thrombospondin 4 |
| chr2_-_40452046 | 1.90 |
ENST00000406785.6
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr3_-_186362223 | 1.87 |
ENST00000265022.8
|
DGKG
|
diacylglycerol kinase gamma |
| chr2_+_156436423 | 1.87 |
ENST00000540309.5
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr5_+_10564064 | 1.85 |
ENST00000296657.7
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr17_+_58755821 | 1.83 |
ENST00000308249.4
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent 1E |
| chr2_+_161416273 | 1.82 |
ENST00000389554.8
|
TBR1
|
T-box brain transcription factor 1 |
| chr1_-_46668454 | 1.82 |
ENST00000576409.5
|
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr11_-_45665578 | 1.81 |
ENST00000308064.7
|
CHST1
|
carbohydrate sulfotransferase 1 |
| chr15_-_79090760 | 1.80 |
ENST00000419573.7
ENST00000558480.7 |
RASGRF1
|
Ras protein specific guanine nucleotide releasing factor 1 |
| chr1_-_31239880 | 1.77 |
ENST00000373736.7
|
NKAIN1
|
sodium/potassium transporting ATPase interacting 1 |
| chr1_-_147172456 | 1.75 |
ENST00000254101.4
|
PRKAB2
|
protein kinase AMP-activated non-catalytic subunit beta 2 |
| chr2_-_36966503 | 1.74 |
ENST00000263918.9
|
STRN
|
striatin |
| chrX_+_111096136 | 1.74 |
ENST00000372007.10
|
PAK3
|
p21 (RAC1) activated kinase 3 |
| chr12_+_109052564 | 1.73 |
ENST00000257548.10
ENST00000536723.5 ENST00000536393.5 |
USP30
|
ubiquitin specific peptidase 30 |
| chr2_-_212538766 | 1.73 |
ENST00000342788.9
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4 |
| chr1_-_211134135 | 1.72 |
ENST00000638983.1
ENST00000271751.10 ENST00000639952.1 |
ENSG00000284299.1
KCNH1
|
novel protein potassium voltage-gated channel subfamily H member 1 |
| chr1_+_231528541 | 1.70 |
ENST00000413309.3
ENST00000366639.9 |
TSNAX
|
translin associated factor X |
| chr10_+_123666355 | 1.65 |
ENST00000284674.2
|
GPR26
|
G protein-coupled receptor 26 |
| chr8_-_52409743 | 1.65 |
ENST00000276480.11
|
ST18
|
ST18 C2H2C-type zinc finger transcription factor |
| chr9_-_10612966 | 1.64 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D |
| chr1_-_217089627 | 1.64 |
ENST00000361525.7
|
ESRRG
|
estrogen related receptor gamma |
| chr12_+_51662781 | 1.63 |
ENST00000355133.7
ENST00000599343.5 |
SCN8A
|
sodium voltage-gated channel alpha subunit 8 |
| chr9_-_109167159 | 1.63 |
ENST00000561981.5
|
FRRS1L
|
ferric chelate reductase 1 like |
| chr7_-_122886706 | 1.61 |
ENST00000313070.11
ENST00000334010.11 ENST00000615869.4 |
CADPS2
|
calcium dependent secretion activator 2 |
| chr10_+_114043858 | 1.60 |
ENST00000369295.4
|
ADRB1
|
adrenoceptor beta 1 |
| chr20_-_44810539 | 1.59 |
ENST00000372851.8
|
RIMS4
|
regulating synaptic membrane exocytosis 4 |
| chr16_-_2214776 | 1.58 |
ENST00000333503.8
|
PGP
|
phosphoglycolate phosphatase |
| chr7_-_139777986 | 1.55 |
ENST00000406875.8
|
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr19_-_409134 | 1.55 |
ENST00000332235.8
|
C2CD4C
|
C2 calcium dependent domain containing 4C |
| chr6_-_11044275 | 1.53 |
ENST00000354666.4
|
ELOVL2
|
ELOVL fatty acid elongase 2 |
| chr9_-_131270493 | 1.52 |
ENST00000372269.7
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78 member A |
| chr22_+_50343294 | 1.52 |
ENST00000359139.7
ENST00000395741.7 ENST00000612753.5 ENST00000395744.7 |
PPP6R2
|
protein phosphatase 6 regulatory subunit 2 |
| chr17_-_7929793 | 1.51 |
ENST00000303790.3
|
KCNAB3
|
potassium voltage-gated channel subfamily A regulatory beta subunit 3 |
| chr16_+_28292485 | 1.49 |
ENST00000341901.5
|
SBK1
|
SH3 domain binding kinase 1 |
| chr12_+_78864768 | 1.48 |
ENST00000261205.9
ENST00000457153.6 |
SYT1
|
synaptotagmin 1 |
| chr5_+_153490655 | 1.47 |
ENST00000518142.5
ENST00000285900.10 |
GRIA1
|
glutamate ionotropic receptor AMPA type subunit 1 |
| chr7_-_44490609 | 1.47 |
ENST00000355451.8
|
NUDCD3
|
NudC domain containing 3 |
| chr12_-_31591129 | 1.45 |
ENST00000389082.10
|
DENND5B
|
DENN domain containing 5B |
| chr1_+_213987929 | 1.45 |
ENST00000498508.6
ENST00000366958.9 |
PROX1
|
prospero homeobox 1 |
| chr8_-_138497254 | 1.44 |
ENST00000395297.6
|
FAM135B
|
family with sequence similarity 135 member B |
| chr11_-_117797091 | 1.42 |
ENST00000527706.5
ENST00000651296.2 ENST00000651172.1 ENST00000321322.6 |
DSCAML1
|
DS cell adhesion molecule like 1 |
| chr16_-_31010611 | 1.42 |
ENST00000215095.11
|
STX1B
|
syntaxin 1B |
| chr5_-_134632769 | 1.40 |
ENST00000505758.5
ENST00000439578.5 ENST00000502286.1 ENST00000402673.7 |
SAR1B
|
secretion associated Ras related GTPase 1B |
| chr20_-_5950463 | 1.39 |
ENST00000203001.7
ENST00000453074.6 |
TRMT6
|
tRNA methyltransferase 6 |
| chr9_-_28670285 | 1.38 |
ENST00000379992.6
ENST00000308675.5 ENST00000613945.3 |
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr21_-_32727933 | 1.35 |
ENST00000357345.7
ENST00000429236.5 |
SYNJ1
|
synaptojanin 1 |
| chr19_+_46601296 | 1.35 |
ENST00000598871.5
ENST00000291295.14 ENST00000594523.5 |
CALM3
|
calmodulin 3 |
| chr2_-_179861604 | 1.35 |
ENST00000410066.7
|
ZNF385B
|
zinc finger protein 385B |
| chr14_+_52552830 | 1.34 |
ENST00000321662.11
|
GPR137C
|
G protein-coupled receptor 137C |
| chr21_+_43865200 | 1.32 |
ENST00000291572.13
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr20_-_57266606 | 1.31 |
ENST00000450594.6
ENST00000395863.8 |
BMP7
|
bone morphogenetic protein 7 |
| chr19_+_51311638 | 1.31 |
ENST00000270642.9
|
IGLON5
|
IgLON family member 5 |
| chr13_-_109786567 | 1.30 |
ENST00000375856.5
|
IRS2
|
insulin receptor substrate 2 |
| chr6_+_117265550 | 1.30 |
ENST00000352536.7
ENST00000326274.6 |
VGLL2
|
vestigial like family member 2 |
| chr1_+_99264473 | 1.29 |
ENST00000370185.9
|
PLPPR4
|
phospholipid phosphatase related 4 |
| chrX_-_40735476 | 1.28 |
ENST00000324817.6
|
MED14
|
mediator complex subunit 14 |
| chr5_-_131796921 | 1.28 |
ENST00000307968.11
ENST00000307954.12 |
FNIP1
|
folliculin interacting protein 1 |
| chr9_+_34990250 | 1.28 |
ENST00000454002.6
ENST00000545841.5 |
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr14_-_55411817 | 1.27 |
ENST00000247178.6
|
ATG14
|
autophagy related 14 |
| chr11_-_63917129 | 1.27 |
ENST00000301459.5
|
RCOR2
|
REST corepressor 2 |
| chr17_-_39607876 | 1.26 |
ENST00000302584.5
|
NEUROD2
|
neuronal differentiation 2 |
| chr19_+_35030438 | 1.26 |
ENST00000415950.5
ENST00000262631.11 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr6_+_68635273 | 1.26 |
ENST00000370598.6
|
ADGRB3
|
adhesion G protein-coupled receptor B3 |
| chr3_-_15859771 | 1.26 |
ENST00000399451.6
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr18_+_69400852 | 1.26 |
ENST00000382713.10
|
DOK6
|
docking protein 6 |
| chr12_-_110583305 | 1.25 |
ENST00000354300.5
|
PPTC7
|
protein phosphatase targeting COQ7 |
| chr17_+_57256514 | 1.25 |
ENST00000284073.7
ENST00000674964.1 |
MSI2
|
musashi RNA binding protein 2 |
| chr17_-_64263221 | 1.24 |
ENST00000258991.7
ENST00000583738.1 ENST00000584379.6 |
TEX2
|
testis expressed 2 |
| chr20_-_44311142 | 1.24 |
ENST00000396825.4
|
FITM2
|
fat storage inducing transmembrane protein 2 |
| chr11_+_64306227 | 1.24 |
ENST00000405666.5
ENST00000468670.2 |
ESRRA
|
estrogen related receptor alpha |
| chr4_-_141133436 | 1.23 |
ENST00000306799.7
ENST00000515673.7 |
RNF150
|
ring finger protein 150 |
| chr22_-_44312894 | 1.22 |
ENST00000381176.5
|
SHISAL1
|
shisa like 1 |
| chr9_-_136050502 | 1.22 |
ENST00000371753.5
|
NACC2
|
NACC family member 2 |
| chr10_+_26216766 | 1.22 |
ENST00000376261.8
|
GAD2
|
glutamate decarboxylase 2 |
| chr11_-_1572261 | 1.22 |
ENST00000397374.8
|
DUSP8
|
dual specificity phosphatase 8 |
| chr8_+_106657836 | 1.20 |
ENST00000312046.10
|
OXR1
|
oxidation resistance 1 |
| chr4_-_86360071 | 1.20 |
ENST00000641677.1
ENST00000639234.1 ENST00000641553.1 ENST00000641826.1 ENST00000641537.1 ENST00000395169.9 ENST00000641408.1 ENST00000638225.1 ENST00000641052.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr8_-_104588998 | 1.19 |
ENST00000424843.6
|
LRP12
|
LDL receptor related protein 12 |
| chr14_+_102362931 | 1.19 |
ENST00000359520.12
|
TECPR2
|
tectonin beta-propeller repeat containing 2 |
| chr1_-_84690406 | 1.19 |
ENST00000605755.5
ENST00000342203.8 ENST00000437941.6 |
SSX2IP
|
SSX family member 2 interacting protein |
| chr4_+_41360759 | 1.19 |
ENST00000508501.5
ENST00000512946.5 ENST00000313860.11 ENST00000512632.5 ENST00000512820.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr10_-_67696115 | 1.18 |
ENST00000433211.7
|
CTNNA3
|
catenin alpha 3 |
| chr7_+_28412511 | 1.17 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr13_+_34942263 | 1.17 |
ENST00000379939.7
ENST00000400445.7 |
NBEA
|
neurobeachin |
| chr1_+_15617415 | 1.17 |
ENST00000480945.6
|
DDI2
|
DNA damage inducible 1 homolog 2 |
| chr15_-_61229297 | 1.15 |
ENST00000335670.11
|
RORA
|
RAR related orphan receptor A |
| chr4_+_150078426 | 1.15 |
ENST00000296550.12
|
DCLK2
|
doublecortin like kinase 2 |
| chr6_+_96015964 | 1.15 |
ENST00000302103.6
|
FUT9
|
fucosyltransferase 9 |
| chr8_+_141391989 | 1.14 |
ENST00000520105.5
ENST00000523147.5 ENST00000521578.6 |
PTP4A3
|
protein tyrosine phosphatase 4A3 |
| chr1_+_220528112 | 1.14 |
ENST00000366917.6
ENST00000402574.5 ENST00000611084.4 ENST00000366918.8 |
MARK1
|
microtubule affinity regulating kinase 1 |
| chr10_-_689613 | 1.12 |
ENST00000280886.12
ENST00000634311.1 |
DIP2C
|
disco interacting protein 2 homolog C |
| chrX_+_16946650 | 1.12 |
ENST00000357277.8
|
REPS2
|
RALBP1 associated Eps domain containing 2 |
| chr5_-_59893718 | 1.12 |
ENST00000340635.11
|
PDE4D
|
phosphodiesterase 4D |
| chr22_-_33058368 | 1.11 |
ENST00000358763.7
|
SYN3
|
synapsin III |
| chr6_-_13711817 | 1.10 |
ENST00000011619.6
|
RANBP9
|
RAN binding protein 9 |
| chrX_-_46759055 | 1.09 |
ENST00000328306.4
ENST00000616978.5 |
SLC9A7
|
solute carrier family 9 member A7 |
| chr1_-_40665654 | 1.09 |
ENST00000372684.8
|
RIMS3
|
regulating synaptic membrane exocytosis 3 |
| chr14_+_79279403 | 1.08 |
ENST00000281127.11
|
NRXN3
|
neurexin 3 |
| chr5_-_58460076 | 1.07 |
ENST00000274289.8
ENST00000617412.1 |
PLK2
|
polo like kinase 2 |
| chr9_-_14693419 | 1.06 |
ENST00000380916.9
|
ZDHHC21
|
zinc finger DHHC-type palmitoyltransferase 21 |
| chr7_+_111091006 | 1.06 |
ENST00000451085.5
ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr19_+_13024573 | 1.05 |
ENST00000358552.7
ENST00000360105.8 ENST00000588228.5 ENST00000676441.1 ENST00000591028.1 |
NFIX
|
nuclear factor I X |
| chr2_+_165239388 | 1.05 |
ENST00000424833.5
ENST00000375437.7 ENST00000631182.3 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr16_+_12901591 | 1.05 |
ENST00000558583.3
|
SHISA9
|
shisa family member 9 |
| chr15_+_33310946 | 1.04 |
ENST00000415757.7
ENST00000634891.2 ENST00000389232.9 ENST00000622037.1 |
RYR3
|
ryanodine receptor 3 |
| chr12_-_10723307 | 1.04 |
ENST00000279550.11
ENST00000228251.9 |
YBX3
|
Y-box binding protein 3 |
| chr1_+_42380772 | 1.03 |
ENST00000431473.4
ENST00000410070.6 |
RIMKLA
|
ribosomal modification protein rimK like family member A |
| chr11_+_17734732 | 1.03 |
ENST00000379472.4
ENST00000675775.1 ENST00000265969.8 ENST00000640318.2 ENST00000639325.2 |
KCNC1
|
potassium voltage-gated channel subfamily C member 1 |
| chr3_+_20040437 | 1.03 |
ENST00000263754.5
|
KAT2B
|
lysine acetyltransferase 2B |
| chr16_+_11668414 | 1.02 |
ENST00000329565.6
|
SNN
|
stannin |
| chr7_+_136868622 | 1.02 |
ENST00000680005.1
ENST00000445907.6 |
CHRM2
|
cholinergic receptor muscarinic 2 |
| chrX_+_114584037 | 1.02 |
ENST00000371951.5
ENST00000371950.3 ENST00000276198.6 |
HTR2C
|
5-hydroxytryptamine receptor 2C |
| chr3_-_24494791 | 1.02 |
ENST00000431815.5
ENST00000356447.9 ENST00000418247.1 ENST00000416420.5 ENST00000396671.7 |
THRB
|
thyroid hormone receptor beta |
| chr10_+_132537778 | 1.02 |
ENST00000368594.8
|
INPP5A
|
inositol polyphosphate-5-phosphatase A |
| chr6_-_55579178 | 1.02 |
ENST00000308161.8
ENST00000398661.6 ENST00000274901.9 |
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase like 1 |
| chr18_+_13218769 | 1.01 |
ENST00000677055.1
ENST00000399848.7 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr8_+_131904071 | 1.01 |
ENST00000254624.10
ENST00000522709.5 |
EFR3A
|
EFR3 homolog A |
| chr4_-_104494882 | 1.00 |
ENST00000394767.3
|
CXXC4
|
CXXC finger protein 4 |
| chr1_-_72282457 | 1.00 |
ENST00000357731.10
|
NEGR1
|
neuronal growth regulator 1 |
| chr17_+_12020812 | 1.00 |
ENST00000415385.7
ENST00000353533.10 |
MAP2K4
|
mitogen-activated protein kinase kinase 4 |
| chr1_-_38005484 | 1.00 |
ENST00000373016.4
|
FHL3
|
four and a half LIM domains 3 |
| chr8_+_38176802 | 0.96 |
ENST00000287322.5
|
BAG4
|
BAG cochaperone 4 |
| chr14_-_44961889 | 0.94 |
ENST00000579157.1
ENST00000396128.9 ENST00000556500.1 |
KLHL28
|
kelch like family member 28 |
| chr11_-_73598183 | 0.94 |
ENST00000064778.8
|
FAM168A
|
family with sequence similarity 168 member A |
| chr1_-_155562693 | 0.93 |
ENST00000368346.7
ENST00000392403.8 ENST00000679333.1 ENST00000679133.1 |
ASH1L
|
ASH1 like histone lysine methyltransferase |
| chr7_+_98106852 | 0.93 |
ENST00000297293.6
|
LMTK2
|
lemur tyrosine kinase 2 |
| chr8_-_102238903 | 0.93 |
ENST00000251810.8
|
RRM2B
|
ribonucleotide reductase regulatory TP53 inducible subunit M2B |
| chr7_-_105388881 | 0.91 |
ENST00000460391.5
ENST00000393651.8 |
SRPK2
|
SRSF protein kinase 2 |
| chr2_+_197515565 | 0.91 |
ENST00000233892.8
ENST00000409916.5 |
MOB4
|
MOB family member 4, phocein |
| chr4_+_92303946 | 0.91 |
ENST00000282020.9
|
GRID2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr2_-_73233206 | 0.91 |
ENST00000258083.3
|
PRADC1
|
protease associated domain containing 1 |
| chr14_+_93430853 | 0.91 |
ENST00000553484.5
|
UNC79
|
unc-79 homolog, NALCN channel complex subunit |
| chr3_-_120094436 | 0.91 |
ENST00000264235.13
ENST00000677034.1 |
GSK3B
|
glycogen synthase kinase 3 beta |
| chr3_-_126357399 | 0.90 |
ENST00000296233.4
|
KLF15
|
Kruppel like factor 15 |
| chr2_+_86720282 | 0.90 |
ENST00000283632.5
|
RMND5A
|
required for meiotic nuclear division 5 homolog A |
| chr2_-_224585354 | 0.90 |
ENST00000264414.9
ENST00000344951.8 |
CUL3
|
cullin 3 |
| chr16_-_29899043 | 0.89 |
ENST00000346932.9
ENST00000350527.7 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog like 2 |
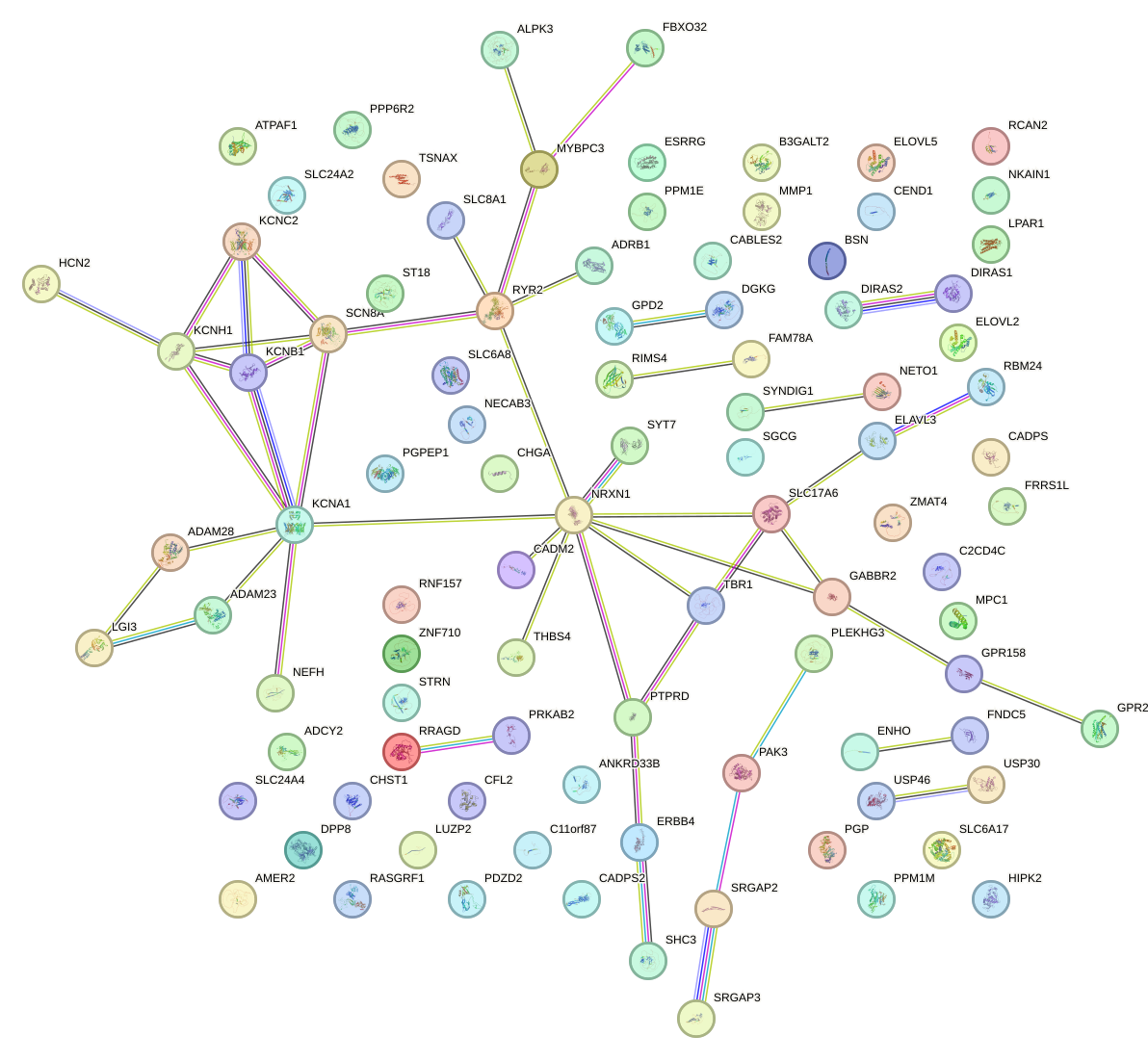
Network of associatons between targets according to the STRING database.
First level regulatory network of UAUUGCU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 1.0 | 3.0 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 1.0 | 2.9 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.9 | 2.7 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.8 | 4.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.8 | 2.4 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.8 | 2.3 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.7 | 1.4 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.7 | 2.0 | GO:0015881 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.6 | 2.4 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.5 | 1.6 | GO:0003099 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.5 | 2.0 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.5 | 1.9 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.5 | 1.4 | GO:0090425 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.5 | 0.5 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.4 | 1.7 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.4 | 2.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.4 | 3.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.4 | 3.3 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.4 | 1.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.4 | 0.8 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.4 | 1.9 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.3 | 4.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 1.7 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.3 | 5.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.3 | 1.0 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.3 | 1.4 | GO:1904980 | positive regulation of endosome organization(GO:1904980) |
| 0.3 | 2.0 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.3 | 1.3 | GO:1905069 | nephrogenic mesenchyme morphogenesis(GO:0072134) allantois development(GO:1905069) |
| 0.3 | 1.0 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.3 | 1.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.3 | 0.9 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.3 | 1.9 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.3 | 1.8 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.3 | 1.5 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.3 | 1.7 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.3 | 2.0 | GO:0021764 | amygdala development(GO:0021764) |
| 0.3 | 2.0 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.3 | 0.8 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.3 | 2.7 | GO:1990253 | cellular response to leucine(GO:0071233) cellular response to leucine starvation(GO:1990253) |
| 0.3 | 0.8 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.3 | 1.3 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.3 | 0.8 | GO:1901383 | negative regulation of chorionic trophoblast cell proliferation(GO:1901383) |
| 0.2 | 1.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 4.1 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.2 | 0.7 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.2 | 1.6 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 2.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 4.2 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 2.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.2 | 1.3 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.2 | 3.4 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.2 | 2.7 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 1.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.2 | 1.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 | 0.4 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.2 | 2.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 2.6 | GO:0015820 | leucine transport(GO:0015820) |
| 0.2 | 2.7 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 0.6 | GO:0045175 | basal protein localization(GO:0045175) |
| 0.2 | 0.8 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.2 | 3.7 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.2 | 1.3 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 1.2 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.2 | 2.3 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.2 | 0.6 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.2 | 1.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 1.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 0.9 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.2 | 1.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 0.9 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.5 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 1.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.6 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 1.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.4 | GO:2000559 | CD24 biosynthetic process(GO:0035724) activation of meiosis involved in egg activation(GO:0060466) negative regulation of monocyte extravasation(GO:2000438) regulation of CD24 biosynthetic process(GO:2000559) positive regulation of CD24 biosynthetic process(GO:2000560) |
| 0.1 | 2.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 2.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 2.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.9 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.1 | 0.5 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.1 | 0.5 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.4 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.2 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.1 | 1.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 2.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 2.2 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.1 | 1.2 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 1.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.9 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.3 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 0.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 2.3 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 1.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.9 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 1.4 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.3 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 1.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 1.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.6 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.7 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.5 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 1.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.9 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 1.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.3 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 1.8 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.4 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 3.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 0.9 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.2 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 1.1 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 1.1 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 1.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 3.8 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.7 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 1.8 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 1.4 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 1.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 1.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 2.7 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 2.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 1.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 0.6 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.9 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.7 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 1.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.8 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 2.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 1.7 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 2.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 1.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 2.7 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 1.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.7 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 1.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 3.0 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 2.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.3 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.8 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 1.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.6 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.5 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.3 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 2.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 1.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 1.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.3 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 1.1 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 1.0 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 1.4 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 2.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.3 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.1 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.2 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 1.2 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 1.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 2.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.7 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0099569 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.8 | 3.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.7 | 3.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.5 | 2.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.4 | 2.7 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 4.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.4 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.3 | 2.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 1.3 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.3 | 0.9 | GO:0034657 | GID complex(GO:0034657) |
| 0.3 | 2.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 1.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.5 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 4.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 1.8 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 4.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 6.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 2.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.2 | 0.8 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.2 | 4.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 2.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 6.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 1.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 1.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 0.5 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.2 | 2.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.6 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 2.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 3.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 5.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.6 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 13.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.9 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 1.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 2.0 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.8 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 6.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 3.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 3.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.6 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 4.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.8 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 3.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.7 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 1.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 3.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 2.5 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 1.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.3 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.9 | 4.7 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.7 | 2.0 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.6 | 3.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.5 | 1.6 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.4 | 1.6 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.4 | 1.9 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.3 | 2.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 3.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.3 | 2.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.3 | 10.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 0.8 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.3 | 2.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.3 | 1.0 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.3 | 0.8 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.3 | 1.3 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.3 | 0.8 | GO:0038100 | nodal binding(GO:0038100) |
| 0.3 | 5.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 1.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 2.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 0.5 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 1.9 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.2 | 4.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 1.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 2.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 2.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.8 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 1.6 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.2 | 1.4 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 1.5 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 1.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 1.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 1.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.9 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 1.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 1.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 1.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.6 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.1 | 3.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.9 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.7 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 1.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.4 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.1 | 1.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.5 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.2 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 3.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 3.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.5 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 4.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 1.0 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 2.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.5 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 1.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.2 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 1.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.7 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 1.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 2.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 2.3 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 1.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 6.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 0.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 2.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 1.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 2.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 6.2 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 0.9 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 1.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 1.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 1.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 2.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 2.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 6.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.8 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 1.4 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 4.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 3.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.4 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 2.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.8 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.9 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.5 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 1.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 4.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 2.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 1.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 3.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 2.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 1.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.0 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 1.7 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 4.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 5.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 4.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 3.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 2.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 3.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 16.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 4.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 1.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 4.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 11.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 6.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 5.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 3.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 1.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 5.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.5 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.5 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.8 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 2.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |