Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for VAX2_RHOXF2
Z-value: 1.06
Transcription factors associated with VAX2_RHOXF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
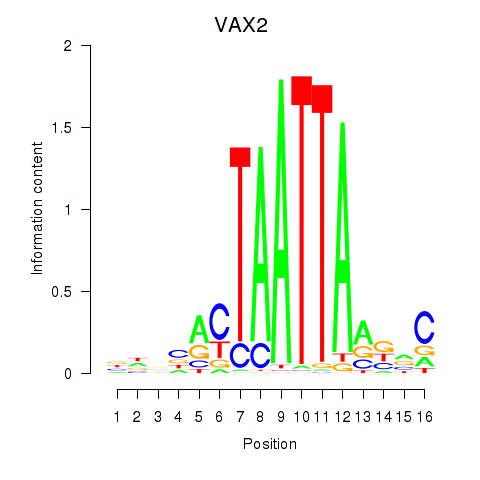
VAX2
|
ENSG00000116035.4 | ventral anterior homeobox 2 |
|
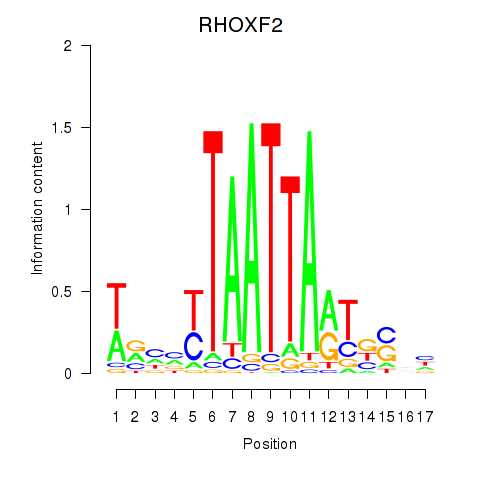
RHOXF2
|
ENSG00000131721.6 | Rhox homeobox family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RHOXF2 | hg38_v1_chrX_+_120158600_120158624 | -0.51 | 3.0e-03 | Click! |
| VAX2 | hg38_v1_chr2_+_70900546_70900617 | 0.15 | 4.3e-01 | Click! |
Activity profile of VAX2_RHOXF2 motif
Sorted Z-values of VAX2_RHOXF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_122370523 | 4.18 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr9_+_122371014 | 4.15 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr9_+_122371036 | 4.10 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr16_+_72054477 | 4.09 |
ENST00000355906.10
ENST00000570083.5 ENST00000228226.12 ENST00000398131.6 ENST00000569639.5 ENST00000564499.5 ENST00000357763.8 ENST00000613898.1 ENST00000562526.5 ENST00000565574.5 ENST00000568417.6 |
HP
|
haptoglobin |
| chr16_-_55833085 | 3.07 |
ENST00000360526.8
|
CES1
|
carboxylesterase 1 |
| chr16_-_55833186 | 3.07 |
ENST00000361503.8
ENST00000422046.6 |
CES1
|
carboxylesterase 1 |
| chr20_+_57561103 | 2.79 |
ENST00000319441.6
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 |
| chr2_-_88947820 | 2.56 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr6_+_26365159 | 2.55 |
ENST00000532865.5
ENST00000396934.7 ENST00000508906.6 ENST00000530653.5 ENST00000527417.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr6_+_26365215 | 2.48 |
ENST00000527422.5
ENST00000356386.6 ENST00000396948.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr4_-_185775271 | 2.38 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_26365176 | 2.19 |
ENST00000377708.7
|
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr2_+_90038848 | 2.13 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr5_+_132873660 | 2.09 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chr6_+_29550407 | 2.07 |
ENST00000641137.1
|
OR2I1P
|
olfactory receptor family 2 subfamily I member 1 pseudogene |
| chr3_+_157436842 | 1.91 |
ENST00000295927.4
|
PTX3
|
pentraxin 3 |
| chr2_-_89177160 | 1.91 |
ENST00000484817.1
|
IGKV2-24
|
immunoglobulin kappa variable 2-24 |
| chr4_-_185775411 | 1.87 |
ENST00000445115.5
ENST00000451701.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_26402237 | 1.85 |
ENST00000476549.6
ENST00000450085.6 ENST00000425234.6 ENST00000427334.5 ENST00000506698.1 ENST00000289361.11 |
BTN3A1
|
butyrophilin subfamily 3 member A1 |
| chr6_+_26402289 | 1.80 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin subfamily 3 member A1 |
| chr4_+_168497044 | 1.63 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_+_168497066 | 1.63 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_+_150582119 | 1.60 |
ENST00000317605.6
|
MAB21L2
|
mab-21 like 2 |
| chr9_-_92404559 | 1.57 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chr1_+_162366896 | 1.56 |
ENST00000420220.1
|
ENSG00000254706.3
|
novel protein |
| chr6_+_130018565 | 1.42 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr2_+_90220727 | 1.42 |
ENST00000471857.2
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr4_-_25863537 | 1.36 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr19_+_49513353 | 1.33 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr17_-_42185452 | 1.31 |
ENST00000293330.1
|
HCRT
|
hypocretin neuropeptide precursor |
| chr16_+_72063226 | 1.20 |
ENST00000540303.7
ENST00000356967.6 ENST00000561690.1 |
HPR
|
haptoglobin-related protein |
| chr1_-_92486916 | 1.18 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr4_-_185775432 | 1.18 |
ENST00000457247.5
ENST00000435480.5 ENST00000425679.5 ENST00000457934.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_185775376 | 1.17 |
ENST00000456596.5
ENST00000414724.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_106360668 | 1.15 |
ENST00000633556.3
|
CRYBG1
|
crystallin beta-gamma domain containing 1 |
| chr7_+_116222804 | 1.13 |
ENST00000393481.6
|
TES
|
testin LIM domain protein |
| chr12_+_16347102 | 1.08 |
ENST00000536371.5
ENST00000010404.6 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr12_+_16347665 | 1.04 |
ENST00000535309.5
ENST00000540056.5 ENST00000396209.5 ENST00000540126.5 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr4_+_87975667 | 1.03 |
ENST00000237623.11
ENST00000682655.1 ENST00000508233.6 ENST00000360804.4 ENST00000395080.8 |
SPP1
|
secreted phosphoprotein 1 |
| chr1_+_34792990 | 1.03 |
ENST00000450137.1
ENST00000342280.5 |
GJA4
|
gap junction protein alpha 4 |
| chr9_-_19149278 | 1.01 |
ENST00000434144.5
|
PLIN2
|
perilipin 2 |
| chr10_+_72893734 | 1.00 |
ENST00000334011.10
|
OIT3
|
oncoprotein induced transcript 3 |
| chr14_+_22096017 | 1.00 |
ENST00000390452.2
|
TRDV1
|
T cell receptor delta variable 1 |
| chr12_+_130953898 | 0.99 |
ENST00000261654.10
|
ADGRD1
|
adhesion G protein-coupled receptor D1 |
| chr5_+_172641241 | 0.98 |
ENST00000369800.6
ENST00000520919.5 ENST00000522853.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr12_+_16347637 | 0.96 |
ENST00000543076.5
ENST00000396210.8 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr4_-_73620391 | 0.94 |
ENST00000395777.6
ENST00000307439.10 |
RASSF6
|
Ras association domain family member 6 |
| chr1_-_113871665 | 0.91 |
ENST00000528414.5
ENST00000460620.5 ENST00000359785.10 ENST00000420377.6 ENST00000525799.1 ENST00000538253.5 |
PTPN22
|
protein tyrosine phosphatase non-receptor type 22 |
| chrX_+_136648138 | 0.89 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr8_-_100649660 | 0.85 |
ENST00000311812.7
|
SNX31
|
sorting nexin 31 |
| chr6_-_132714045 | 0.81 |
ENST00000367928.5
|
VNN1
|
vanin 1 |
| chr8_-_33567118 | 0.79 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122 |
| chr7_+_73830988 | 0.78 |
ENST00000340958.4
|
CLDN4
|
claudin 4 |
| chr7_+_100119607 | 0.76 |
ENST00000262932.5
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr4_+_87975829 | 0.75 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1 |
| chr4_-_73620629 | 0.73 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr15_+_58410543 | 0.73 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr20_-_35147285 | 0.71 |
ENST00000374491.3
ENST00000374492.8 |
EDEM2
|
ER degradation enhancing alpha-mannosidase like protein 2 |
| chr12_-_9869345 | 0.70 |
ENST00000228438.3
|
CLEC2B
|
C-type lectin domain family 2 member B |
| chrX_+_136648214 | 0.67 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr16_+_11965193 | 0.67 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr6_+_26440472 | 0.65 |
ENST00000494393.5
ENST00000482451.5 ENST00000471353.5 ENST00000361232.7 ENST00000487627.5 ENST00000496719.1 ENST00000244519.7 ENST00000490254.5 ENST00000487272.1 |
BTN3A3
|
butyrophilin subfamily 3 member A3 |
| chr2_-_189179754 | 0.63 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr15_-_37101205 | 0.63 |
ENST00000338564.9
ENST00000558313.5 ENST00000340545.9 |
MEIS2
|
Meis homeobox 2 |
| chrX_+_43656289 | 0.61 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr7_+_50308672 | 0.60 |
ENST00000439701.2
ENST00000438033.5 ENST00000492782.6 |
IKZF1
|
IKAROS family zinc finger 1 |
| chr21_-_36542600 | 0.60 |
ENST00000399136.5
|
CLDN14
|
claudin 14 |
| chr10_+_18260715 | 0.60 |
ENST00000615785.4
ENST00000617363.4 ENST00000396576.6 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr19_-_45584769 | 0.60 |
ENST00000263275.5
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3 |
| chr2_+_89884740 | 0.59 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr4_-_67963441 | 0.59 |
ENST00000508048.6
|
TMPRSS11A
|
transmembrane serine protease 11A |
| chr3_+_158801926 | 0.58 |
ENST00000622669.4
ENST00000392813.8 ENST00000415822.8 ENST00000651862.1 ENST00000264266.12 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr7_+_142300924 | 0.58 |
ENST00000455382.2
|
TRBV2
|
T cell receptor beta variable 2 |
| chr14_+_21997531 | 0.58 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr10_+_97446137 | 0.56 |
ENST00000370854.7
ENST00000393760.6 ENST00000414567.5 |
ZDHHC16
|
zinc finger DHHC-type palmitoyltransferase 16 |
| chr4_-_137532452 | 0.55 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr2_-_40453438 | 0.55 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr10_+_97446194 | 0.55 |
ENST00000370846.8
ENST00000352634.8 ENST00000353979.7 ENST00000370842.6 ENST00000345745.9 |
ZDHHC16
|
zinc finger DHHC-type palmitoyltransferase 16 |
| chr1_-_56579555 | 0.54 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr15_+_73926443 | 0.53 |
ENST00000261921.8
|
LOXL1
|
lysyl oxidase like 1 |
| chr3_-_151316795 | 0.52 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr5_+_177384430 | 0.48 |
ENST00000512593.5
ENST00000324417.6 |
SLC34A1
|
solute carrier family 34 member 1 |
| chr7_-_44541262 | 0.47 |
ENST00000289547.8
ENST00000546276.5 ENST00000423141.1 |
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr4_-_73620500 | 0.47 |
ENST00000335049.5
|
RASSF6
|
Ras association domain family member 6 |
| chr4_+_40200320 | 0.46 |
ENST00000511121.5
|
RHOH
|
ras homolog family member H |
| chrX_-_155071064 | 0.45 |
ENST00000369484.8
ENST00000369476.8 |
CMC4
MTCP1
|
C-X9-C motif containing 4 mature T cell proliferation 1 |
| chr15_+_49170237 | 0.43 |
ENST00000560031.6
ENST00000558145.5 ENST00000544523.5 ENST00000560138.5 |
GALK2
|
galactokinase 2 |
| chr16_-_28597042 | 0.42 |
ENST00000533150.5
ENST00000335715.9 |
SULT1A2
|
sulfotransferase family 1A member 2 |
| chr4_-_106368772 | 0.41 |
ENST00000638719.4
|
GIMD1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr12_+_64405046 | 0.38 |
ENST00000540203.5
|
XPOT
|
exportin for tRNA |
| chr2_+_90209873 | 0.37 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr15_+_49170200 | 0.36 |
ENST00000396509.6
|
GALK2
|
galactokinase 2 |
| chr11_-_31509569 | 0.35 |
ENST00000526776.5
|
IMMP1L
|
inner mitochondrial membrane peptidase subunit 1 |
| chr4_+_168497113 | 0.35 |
ENST00000511948.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr15_+_58138368 | 0.35 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr2_-_89297785 | 0.34 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr15_+_58138169 | 0.34 |
ENST00000558772.5
|
AQP9
|
aquaporin 9 |
| chr12_-_21910853 | 0.34 |
ENST00000544039.5
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr7_-_100119291 | 0.33 |
ENST00000431404.2
|
TAF6
|
TATA-box binding protein associated factor 6 |
| chr9_-_5339874 | 0.33 |
ENST00000223862.2
|
RLN1
|
relaxin 1 |
| chr4_-_40475749 | 0.33 |
ENST00000507180.5
|
RBM47
|
RNA binding motif protein 47 |
| chr6_-_137173644 | 0.31 |
ENST00000296980.7
ENST00000349184.8 ENST00000339602.3 |
IL22RA2
|
interleukin 22 receptor subunit alpha 2 |
| chr11_-_117876683 | 0.30 |
ENST00000530956.6
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr7_-_44541318 | 0.29 |
ENST00000381160.8
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr7_-_111392915 | 0.29 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr5_-_150449676 | 0.29 |
ENST00000312037.6
|
RPS14
|
ribosomal protein S14 |
| chr12_+_26195647 | 0.28 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr5_-_150449696 | 0.27 |
ENST00000401695.8
|
RPS14
|
ribosomal protein S14 |
| chr8_-_96261579 | 0.27 |
ENST00000517720.1
ENST00000523821.5 ENST00000287025.4 |
MTERF3
|
mitochondrial transcription termination factor 3 |
| chr11_-_117876892 | 0.26 |
ENST00000539526.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr14_+_22202561 | 0.25 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr19_+_24033399 | 0.24 |
ENST00000664647.1
ENST00000656993.1 ENST00000666101.1 ENST00000613065.4 |
ENSG00000268362.6
ZNF254
|
novel transcript zinc finger protein 254 |
| chr12_-_50249883 | 0.24 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr7_-_101217569 | 0.24 |
ENST00000223127.8
|
PLOD3
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 |
| chr6_-_82247697 | 0.23 |
ENST00000306270.12
ENST00000610980.4 |
IBTK
|
inhibitor of Bruton tyrosine kinase |
| chr2_-_213151590 | 0.23 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr2_+_102473219 | 0.22 |
ENST00000295269.5
|
SLC9A4
|
solute carrier family 9 member A4 |
| chrX_+_73563190 | 0.22 |
ENST00000373504.10
ENST00000373502.9 |
CHIC1
|
cysteine rich hydrophobic domain 1 |
| chr11_-_117876719 | 0.21 |
ENST00000529335.6
ENST00000260282.8 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr1_+_160400543 | 0.21 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr17_+_50746534 | 0.21 |
ENST00000511974.5
|
LUC7L3
|
LUC7 like 3 pre-mRNA splicing factor |
| chr5_+_174724549 | 0.21 |
ENST00000239243.7
ENST00000507785.2 |
MSX2
|
msh homeobox 2 |
| chr17_-_41047267 | 0.21 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr19_-_3557563 | 0.20 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr21_-_7793954 | 0.20 |
ENST00000624951.1
|
SMIM34B
|
small integral membrane protein 34B |
| chr12_+_55444069 | 0.19 |
ENST00000641516.1
ENST00000641202.1 |
OR6C2
|
olfactory receptor family 6 subfamily C member 2 |
| chr11_+_124183219 | 0.19 |
ENST00000641351.2
|
OR10D3
|
olfactory receptor family 10 subfamily D member 3 |
| chr4_-_68245683 | 0.18 |
ENST00000332644.6
|
TMPRSS11B
|
transmembrane serine protease 11B |
| chr14_+_61187544 | 0.17 |
ENST00000555185.5
ENST00000557294.5 ENST00000556778.5 |
PRKCH
|
protein kinase C eta |
| chr9_+_72577788 | 0.17 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr9_-_134068012 | 0.17 |
ENST00000303407.12
|
BRD3
|
bromodomain containing 3 |
| chr17_+_74935892 | 0.15 |
ENST00000328801.6
|
OTOP3
|
otopetrin 3 |
| chr9_+_72577369 | 0.15 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr11_-_117877463 | 0.14 |
ENST00000527717.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr9_+_12693327 | 0.14 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr14_-_21526202 | 0.14 |
ENST00000450879.2
|
SALL2
|
spalt like transcription factor 2 |
| chr5_+_67004618 | 0.13 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_+_29099490 | 0.13 |
ENST00000641659.2
|
OR2J1
|
olfactory receptor family 2 subfamily J member 1 |
| chr21_-_34423951 | 0.13 |
ENST00000450895.2
|
SMIM34A
|
small integral membrane protein 34A |
| chr20_-_51802433 | 0.12 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr17_-_40799939 | 0.12 |
ENST00000306658.8
|
KRT28
|
keratin 28 |
| chr3_+_42979281 | 0.12 |
ENST00000488863.5
ENST00000430121.3 |
GASK1A
|
golgi associated kinase 1A |
| chr6_-_138545685 | 0.10 |
ENST00000342260.9
|
NHSL1
|
NHS like 1 |
| chr6_+_49499916 | 0.10 |
ENST00000371197.9
|
GLYATL3
|
glycine-N-acyltransferase like 3 |
| chr10_+_70052582 | 0.10 |
ENST00000676699.1
|
MACROH2A2
|
macroH2A.2 histone |
| chr19_-_51804104 | 0.10 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr11_-_122116215 | 0.10 |
ENST00000560104.2
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr18_-_69956924 | 0.08 |
ENST00000581982.5
ENST00000280200.8 |
CD226
|
CD226 molecule |
| chr5_+_121961955 | 0.08 |
ENST00000339397.5
|
SRFBP1
|
serum response factor binding protein 1 |
| chr18_+_49561013 | 0.07 |
ENST00000583083.1
|
LIPG
|
lipase G, endothelial type |
| chr1_-_17439657 | 0.06 |
ENST00000375436.9
|
RCC2
|
regulator of chromosome condensation 2 |
| chr7_-_44082464 | 0.06 |
ENST00000335195.10
ENST00000395831.7 ENST00000414235.5 ENST00000242248.10 ENST00000452049.1 |
POLM
|
DNA polymerase mu |
| chr14_-_52791597 | 0.05 |
ENST00000216410.8
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr15_+_101812202 | 0.05 |
ENST00000332238.5
|
OR4F15
|
olfactory receptor family 4 subfamily F member 15 |
| chr12_+_119668109 | 0.05 |
ENST00000229328.10
ENST00000630317.1 |
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr14_+_20455210 | 0.04 |
ENST00000557344.5
ENST00000216714.8 ENST00000398030.8 ENST00000557181.5 ENST00000555839.5 ENST00000553368.1 ENST00000556054.5 ENST00000557054.1 ENST00000557592.5 ENST00000557150.5 |
APEX1
|
apurinic/apyrimidinic endodeoxyribonuclease 1 |
| chr12_-_10802627 | 0.04 |
ENST00000240687.2
|
TAS2R7
|
taste 2 receptor member 7 |
| chr18_+_34976928 | 0.04 |
ENST00000591734.5
ENST00000413393.5 ENST00000589180.5 ENST00000587359.5 |
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr10_+_18400562 | 0.03 |
ENST00000377315.5
ENST00000650685.1 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr11_-_62754141 | 0.03 |
ENST00000527994.1
ENST00000394807.5 ENST00000673933.1 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr11_-_5301946 | 0.01 |
ENST00000380224.2
|
OR51B4
|
olfactory receptor family 51 subfamily B member 4 |
| chr6_+_49499965 | 0.01 |
ENST00000545705.1
|
GLYATL3
|
glycine-N-acyltransferase like 3 |
| chr18_-_69956670 | 0.01 |
ENST00000583955.5
|
CD226
|
CD226 molecule |
| chr12_+_77966204 | 0.00 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr8_+_7926337 | 0.00 |
ENST00000400120.3
|
ZNF705B
|
zinc finger protein 705B |
Network of associatons between targets according to the STRING database.
First level regulatory network of VAX2_RHOXF2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 1.4 | 4.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 1.0 | 3.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.9 | 2.8 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.6 | 1.9 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.6 | 12.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 1.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.4 | 1.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.4 | 1.8 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.3 | 11.8 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.2 | 0.7 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.2 | 1.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 1.0 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.2 | 0.5 | GO:2000118 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.2 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.7 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 0.5 | GO:0018277 | protein deamination(GO:0018277) |
| 0.1 | 3.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 1.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 5.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.6 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 1.6 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 1.0 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.1 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.2 | GO:0051795 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) |
| 0.0 | 1.6 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.8 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.6 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 9.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.6 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 1.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.8 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.9 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.8 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 1.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 1.0 | GO:0048265 | response to pain(GO:0048265) |
| 0.0 | 2.1 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.2 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 0.6 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 12.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 1.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 3.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 4.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 6.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 6.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.6 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.2 | 6.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.9 | 2.8 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.8 | 5.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.5 | 1.6 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.3 | 1.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 0.8 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.2 | 1.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 0.8 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 3.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 6.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.7 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.1 | 3.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.6 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 8.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.5 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.5 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 2.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 13.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |