Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
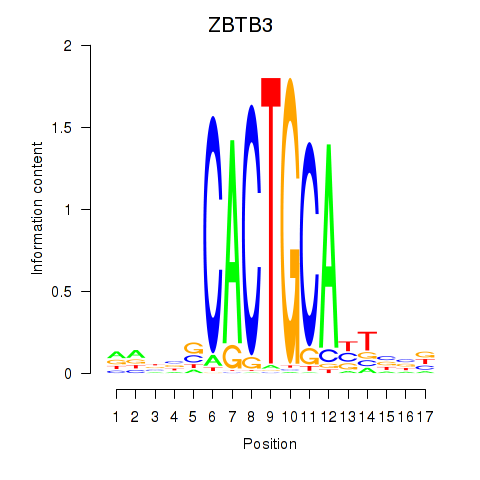
Results for ZBTB3
Z-value: 0.71
Transcription factors associated with ZBTB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB3
|
ENSG00000185670.9 | zinc finger and BTB domain containing 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB3 | hg38_v1_chr11_-_62754141_62754184 | 0.06 | 7.6e-01 | Click! |
Activity profile of ZBTB3 motif
Sorted Z-values of ZBTB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_39281261 | 3.58 |
ENST00000541347.5
ENST00000412814.1 |
CX3CR1
|
C-X3-C motif chemokine receptor 1 |
| chr6_-_41154326 | 2.77 |
ENST00000426005.6
ENST00000437044.2 ENST00000373127.8 |
TREML1
|
triggering receptor expressed on myeloid cells like 1 |
| chr6_+_122779707 | 2.66 |
ENST00000368444.8
ENST00000356535.4 |
FABP7
|
fatty acid binding protein 7 |
| chr12_-_62192762 | 2.36 |
ENST00000416284.8
|
TAFA2
|
TAFA chemokine like family member 2 |
| chr5_-_150290093 | 2.30 |
ENST00000672479.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr21_-_46605073 | 2.24 |
ENST00000291700.9
ENST00000367071.4 |
S100B
|
S100 calcium binding protein B |
| chr14_+_22070548 | 1.51 |
ENST00000390450.3
|
TRAV22
|
T cell receptor alpha variable 22 |
| chr4_+_158201575 | 1.49 |
ENST00000505049.5
ENST00000505189.5 ENST00000511038.5 |
TMEM144
|
transmembrane protein 144 |
| chr2_-_151971750 | 1.45 |
ENST00000636598.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr1_-_110606009 | 1.44 |
ENST00000640774.2
ENST00000638616.2 |
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr1_-_169586539 | 1.43 |
ENST00000367796.3
|
F5
|
coagulation factor V |
| chr12_+_48183602 | 1.35 |
ENST00000316554.5
|
CCDC184
|
coiled-coil domain containing 184 |
| chr1_-_110606347 | 1.31 |
ENST00000316361.10
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr5_+_112738331 | 1.26 |
ENST00000512211.6
|
APC
|
APC regulator of WNT signaling pathway |
| chrX_+_71144818 | 1.24 |
ENST00000536169.5
ENST00000358741.4 ENST00000395855.6 ENST00000374051.7 |
NLGN3
|
neuroligin 3 |
| chr1_-_169586471 | 1.15 |
ENST00000367797.9
|
F5
|
coagulation factor V |
| chr3_-_195442853 | 1.15 |
ENST00000635383.1
ENST00000439666.1 ENST00000618471.4 |
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr18_+_34493289 | 1.14 |
ENST00000682923.1
ENST00000596745.5 ENST00000283365.14 ENST00000315456.10 ENST00000598774.6 ENST00000684266.1 ENST00000683092.1 ENST00000683379.1 ENST00000684359.1 |
DTNA
|
dystrobrevin alpha |
| chr2_-_2326161 | 1.08 |
ENST00000649810.1
ENST00000648318.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr2_-_2326210 | 1.07 |
ENST00000647755.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr5_+_139273752 | 1.06 |
ENST00000509990.5
ENST00000506147.5 ENST00000512107.5 |
MATR3
|
matrin 3 |
| chr19_-_39532809 | 1.06 |
ENST00000326282.5
|
EID2B
|
EP300 interacting inhibitor of differentiation 2B |
| chr6_+_30883085 | 1.01 |
ENST00000504651.5
ENST00000512694.5 ENST00000515233.5 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr6_+_30882914 | 1.00 |
ENST00000509639.5
ENST00000412274.6 ENST00000507901.5 ENST00000507046.5 ENST00000437124.6 ENST00000454612.6 ENST00000396342.6 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr19_+_35030626 | 1.00 |
ENST00000638536.1
|
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr10_-_71773513 | 0.98 |
ENST00000394957.8
|
VSIR
|
V-set immunoregulatory receptor |
| chr7_+_100483919 | 0.97 |
ENST00000300179.7
|
NYAP1
|
neuronal tyrosine phosphorylated phosphoinositide-3-kinase adaptor 1 |
| chr5_-_170297746 | 0.96 |
ENST00000046794.10
|
LCP2
|
lymphocyte cytosolic protein 2 |
| chr1_+_159171607 | 0.95 |
ENST00000368124.8
ENST00000368125.9 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr11_-_83071819 | 0.94 |
ENST00000524635.1
ENST00000526205.5 ENST00000533486.5 ENST00000533276.6 ENST00000527633.6 |
RAB30
|
RAB30, member RAS oncogene family |
| chr1_+_160190567 | 0.94 |
ENST00000368078.8
|
CASQ1
|
calsequestrin 1 |
| chr11_+_26332117 | 0.89 |
ENST00000531646.1
ENST00000256737.8 |
ANO3
|
anoctamin 3 |
| chr19_+_35030438 | 0.89 |
ENST00000415950.5
ENST00000262631.11 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr5_-_95081482 | 0.83 |
ENST00000312216.12
ENST00000512425.5 ENST00000505208.5 ENST00000429576.6 ENST00000508509.5 ENST00000510732.5 |
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr1_-_204494701 | 0.83 |
ENST00000429009.1
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr3_+_98531965 | 0.81 |
ENST00000284311.5
|
GPR15
|
G protein-coupled receptor 15 |
| chr5_+_76083398 | 0.79 |
ENST00000322285.7
|
SV2C
|
synaptic vesicle glycoprotein 2C |
| chr10_-_124744280 | 0.78 |
ENST00000337318.8
|
FAM53B
|
family with sequence similarity 53 member B |
| chr11_-_83071917 | 0.76 |
ENST00000534141.5
|
RAB30
|
RAB30, member RAS oncogene family |
| chr2_-_2326378 | 0.76 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like |
| chrX_+_153072454 | 0.75 |
ENST00000421798.5
|
PNMA6A
|
PNMA family member 6A |
| chr5_+_76083360 | 0.74 |
ENST00000502798.7
|
SV2C
|
synaptic vesicle glycoprotein 2C |
| chr4_-_46388713 | 0.74 |
ENST00000507069.5
|
GABRA2
|
gamma-aminobutyric acid type A receptor subunit alpha2 |
| chr11_+_123358416 | 0.70 |
ENST00000638157.1
|
GRAMD1B
|
GRAM domain containing 1B |
| chr3_-_195442977 | 0.68 |
ENST00000326793.11
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr16_+_67567891 | 0.59 |
ENST00000642847.1
|
CTCF
|
CCCTC-binding factor |
| chr13_-_48037934 | 0.58 |
ENST00000646804.1
ENST00000643246.1 |
SUCLA2
|
succinate-CoA ligase ADP-forming subunit beta |
| chr11_+_64924673 | 0.57 |
ENST00000164133.7
ENST00000532850.1 |
PPP2R5B
|
protein phosphatase 2 regulatory subunit B'beta |
| chr7_-_127585566 | 0.55 |
ENST00000321407.3
|
GCC1
|
GRIP and coiled-coil domain containing 1 |
| chr10_+_68560317 | 0.54 |
ENST00000373644.5
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr1_-_204494752 | 0.53 |
ENST00000684373.1
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr19_-_6670117 | 0.52 |
ENST00000245912.7
|
TNFSF14
|
TNF superfamily member 14 |
| chr12_-_119803383 | 0.52 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr20_+_56412249 | 0.49 |
ENST00000679887.1
ENST00000434344.2 |
CASS4
|
Cas scaffold protein family member 4 |
| chr19_-_48993300 | 0.49 |
ENST00000323798.8
ENST00000263276.6 |
GYS1
|
glycogen synthase 1 |
| chr19_-_55038256 | 0.48 |
ENST00000417454.5
ENST00000310373.7 ENST00000333884.2 |
GP6
|
glycoprotein VI platelet |
| chr12_-_119804298 | 0.46 |
ENST00000678652.1
ENST00000678494.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr19_-_6670151 | 0.45 |
ENST00000675206.1
|
TNFSF14
|
TNF superfamily member 14 |
| chr20_-_63572455 | 0.44 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2 |
| chrX_+_11758159 | 0.44 |
ENST00000361672.6
ENST00000337339.7 ENST00000647869.1 ENST00000312196.10 ENST00000647857.1 ENST00000649130.1 |
MSL3
|
MSL complex subunit 3 |
| chr20_+_56412112 | 0.43 |
ENST00000360314.7
|
CASS4
|
Cas scaffold protein family member 4 |
| chr8_-_19757924 | 0.43 |
ENST00000523262.5
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr11_-_130916437 | 0.41 |
ENST00000533214.1
ENST00000528555.5 ENST00000530356.5 ENST00000265909.9 |
SNX19
|
sorting nexin 19 |
| chr4_-_73069696 | 0.40 |
ENST00000507544.3
ENST00000295890.8 |
COX18
|
cytochrome c oxidase assembly factor COX18 |
| chr9_+_71911615 | 0.37 |
ENST00000334731.7
ENST00000486911.2 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr16_-_68372258 | 0.37 |
ENST00000568373.5
ENST00000563226.1 |
SMPD3
|
sphingomyelin phosphodiesterase 3 |
| chr17_+_77127865 | 0.36 |
ENST00000586429.5
|
SEC14L1
|
SEC14 like lipid binding 1 |
| chr7_-_55538653 | 0.36 |
ENST00000625836.2
|
VOPP1
|
VOPP1 WW domain binding protein |
| chr16_-_19884828 | 0.36 |
ENST00000300571.7
ENST00000570142.5 ENST00000562469.5 |
GPRC5B
|
G protein-coupled receptor class C group 5 member B |
| chr18_+_12407896 | 0.35 |
ENST00000590956.5
ENST00000336990.8 ENST00000440960.6 ENST00000588729.5 |
PRELID3A
|
PRELI domain containing 3A |
| chr8_-_42768602 | 0.33 |
ENST00000534622.5
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit |
| chr5_+_75512058 | 0.32 |
ENST00000514296.5
|
POLK
|
DNA polymerase kappa |
| chr1_+_209827964 | 0.32 |
ENST00000491415.7
|
UTP25
|
UTP25 small subunit processor component |
| chr16_+_89720980 | 0.30 |
ENST00000289816.9
ENST00000568064.5 |
ZNF276
|
zinc finger protein 276 |
| chr4_+_158315309 | 0.30 |
ENST00000460056.6
|
RXFP1
|
relaxin family peptide receptor 1 |
| chr20_+_56412393 | 0.29 |
ENST00000679529.1
|
CASS4
|
Cas scaffold protein family member 4 |
| chr5_-_75511596 | 0.28 |
ENST00000643158.1
ENST00000261415.12 ENST00000646713.1 ENST00000643773.1 ENST00000645866.1 ENST00000644072.2 ENST00000643780.2 ENST00000645483.1 ENST00000642556.1 ENST00000646511.1 |
CERT1
|
ceramide transporter 1 |
| chr21_+_43789522 | 0.28 |
ENST00000497547.2
|
RRP1
|
ribosomal RNA processing 1 |
| chr4_-_5893075 | 0.28 |
ENST00000324989.12
|
CRMP1
|
collapsin response mediator protein 1 |
| chr7_+_23680130 | 0.28 |
ENST00000409192.7
ENST00000409653.5 ENST00000409994.3 ENST00000344962.9 |
FAM221A
|
family with sequence similarity 221 member A |
| chr16_+_30023198 | 0.28 |
ENST00000681219.1
ENST00000300575.6 |
C16orf92
|
chromosome 16 open reading frame 92 |
| chr17_+_47941506 | 0.26 |
ENST00000583599.6
|
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr19_+_52274443 | 0.26 |
ENST00000600821.5
ENST00000595149.5 ENST00000595000.5 ENST00000593612.1 |
ZNF766
|
zinc finger protein 766 |
| chr19_+_39436149 | 0.25 |
ENST00000594990.5
|
SUPT5H
|
SPT5 homolog, DSIF elongation factor subunit |
| chr6_+_29306626 | 0.24 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr1_+_171781635 | 0.24 |
ENST00000361735.4
ENST00000362019.7 ENST00000367737.9 |
EEF1AKNMT
|
eEF1A lysine and N-terminal methyltransferase |
| chr15_+_32030506 | 0.23 |
ENST00000306901.9
ENST00000636440.1 |
CHRNA7
|
cholinergic receptor nicotinic alpha 7 subunit |
| chr1_-_120051714 | 0.23 |
ENST00000579475.7
|
NOTCH2
|
notch receptor 2 |
| chr1_-_156053780 | 0.23 |
ENST00000368309.4
|
UBQLN4
|
ubiquilin 4 |
| chr10_+_49609095 | 0.22 |
ENST00000339797.5
|
CHAT
|
choline O-acetyltransferase |
| chr9_+_123033660 | 0.21 |
ENST00000616002.3
|
GPR21
|
G protein-coupled receptor 21 |
| chrX_-_57680260 | 0.21 |
ENST00000434992.1
|
NLRP2B
|
NLR family pyrin domain containing 2B |
| chr9_+_104504263 | 0.19 |
ENST00000334726.3
|
OR13F1
|
olfactory receptor family 13 subfamily F member 1 |
| chrX_+_135032346 | 0.18 |
ENST00000257013.9
|
RTL8C
|
retrotransposon Gag like 8C |
| chr8_-_42768781 | 0.18 |
ENST00000276410.7
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit |
| chr17_-_5584448 | 0.18 |
ENST00000269280.8
ENST00000571451.6 ENST00000572272.6 ENST00000613500.4 ENST00000619223.4 ENST00000617618.4 ENST00000345221.7 ENST00000262467.10 |
NLRP1
|
NLR family pyrin domain containing 1 |
| chr2_-_74343397 | 0.18 |
ENST00000394019.6
ENST00000377634.8 ENST00000436454.1 |
SLC4A5
|
solute carrier family 4 member 5 |
| chr16_-_58734299 | 0.18 |
ENST00000245206.10
ENST00000434819.2 |
GOT2
|
glutamic-oxaloacetic transaminase 2 |
| chr5_+_77086682 | 0.17 |
ENST00000643365.1
ENST00000645183.1 ENST00000645374.1 ENST00000647364.1 ENST00000643848.1 ENST00000643603.1 ENST00000645459.1 ENST00000643269.1 ENST00000503969.6 ENST00000646262.1 |
ZBED3-AS1
ENSG00000284762.1
|
ZBED3 antisense RNA 1 phosphodiesterase 8B |
| chr5_-_95081858 | 0.17 |
ENST00000505465.1
|
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr2_-_153478753 | 0.17 |
ENST00000325926.4
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator homolog |
| chr1_-_166166973 | 0.17 |
ENST00000354422.4
|
FAM78B
|
family with sequence similarity 78 member B |
| chr2_-_110212519 | 0.14 |
ENST00000611969.5
|
MTLN
|
mitoregulin |
| chr9_+_71911468 | 0.14 |
ENST00000377031.7
|
C9orf85
|
chromosome 9 open reading frame 85 |
| chr16_-_2135898 | 0.12 |
ENST00000262304.9
ENST00000423118.5 |
PKD1
|
polycystin 1, transient receptor potential channel interacting |
| chr10_-_75109106 | 0.11 |
ENST00000607487.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chrX_+_70234655 | 0.10 |
ENST00000374521.4
|
AWAT1
|
acyl-CoA wax alcohol acyltransferase 1 |
| chr1_-_155300933 | 0.10 |
ENST00000434082.3
|
PKLR
|
pyruvate kinase L/R |
| chr7_+_143935233 | 0.09 |
ENST00000408955.3
|
OR2F2
|
olfactory receptor family 2 subfamily F member 2 |
| chr20_-_56497608 | 0.08 |
ENST00000617620.1
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr2_+_240691851 | 0.07 |
ENST00000429564.1
ENST00000337801.9 |
AQP12A
|
aquaporin 12A |
| chr8_+_38404363 | 0.06 |
ENST00000527175.1
|
LETM2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chr15_+_64136330 | 0.06 |
ENST00000560861.1
|
SNX1
|
sorting nexin 1 |
| chr12_+_27466810 | 0.05 |
ENST00000298876.8
ENST00000416383.5 |
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chrX_-_386900 | 0.04 |
ENST00000390665.9
|
PPP2R3B
|
protein phosphatase 2 regulatory subunit B''beta |
| chr5_-_83673544 | 0.04 |
ENST00000503117.1
ENST00000510978.5 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr5_-_175968280 | 0.03 |
ENST00000513482.1
ENST00000265097.9 ENST00000628318.2 |
THOC3
|
THO complex 3 |
| chr10_-_75109172 | 0.02 |
ENST00000372700.7
ENST00000473072.2 ENST00000491677.6 ENST00000372702.7 |
DUSP13
|
dual specificity phosphatase 13 |
| chr10_-_75109085 | 0.01 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chr14_-_106803221 | 0.01 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr13_+_27424583 | 0.01 |
ENST00000381140.10
|
GTF3A
|
general transcription factor IIIA |
| chr14_-_106579223 | 0.00 |
ENST00000390626.2
|
IGHV5-51
|
immunoglobulin heavy variable 5-51 |
| chr11_+_67240047 | 0.00 |
ENST00000530342.2
|
KDM2A
|
lysine demethylase 2A |
| chr19_-_3772211 | 0.00 |
ENST00000555978.5
ENST00000555633.3 |
RAX2
|
retina and anterior neural fold homeobox 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.4 | 2.6 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.3 | 1.9 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.3 | 1.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.3 | 2.7 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.2 | 1.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 | 0.5 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.6 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.1 | 2.2 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.1 | 1.3 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 2.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.4 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 2.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.6 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.1 | 0.9 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 2.7 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 0.4 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.3 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.1 | 1.8 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 0.2 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.1 | 0.2 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.9 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.6 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 2.9 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.3 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 1.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 1.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 1.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.8 | GO:0072678 | T cell migration(GO:0072678) |
| 0.0 | 0.8 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 4.2 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.4 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 0.9 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 2.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 3.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 1.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 1.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 5.4 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 1.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.4 | 1.9 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.6 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 0.6 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 2.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 2.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 2.7 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 2.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.4 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.3 | GO:0004733 | pyridoxamine-phosphate oxidase activity(GO:0004733) |
| 0.1 | 0.4 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.2 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.7 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 1.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 2.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 1.5 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.2 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 2.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 3.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |