|
chr11_-_111913195
Show fit
|
14.32 |
ENST00000531198.5
ENST00000616970.5
ENST00000527899.6
|
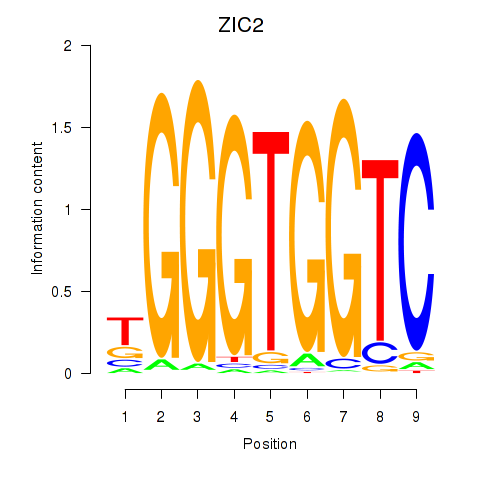
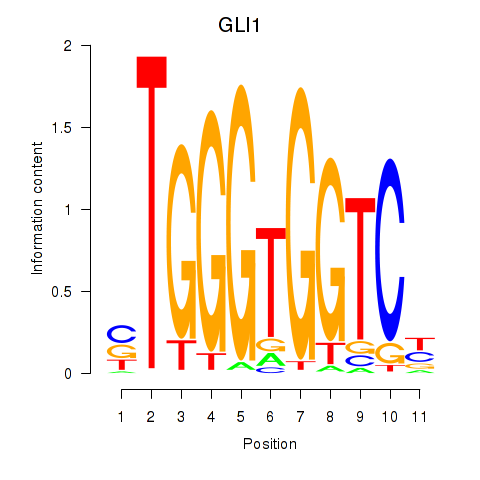
CRYAB
|
crystallin alpha B
|
|
chr11_-_111913134
Show fit
|
12.92 |
ENST00000533971.2
ENST00000526180.6
|
CRYAB
|
crystallin alpha B
|
|
chr19_+_44809089
Show fit
|
12.00 |
ENST00000270233.12
ENST00000591520.6
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr19_+_44809053
Show fit
|
11.81 |
ENST00000611077.5
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr1_+_46798998
Show fit
|
10.30 |
ENST00000640628.1
ENST00000271153.8
ENST00000371923.9
ENST00000371919.8
ENST00000614163.4
|
CYP4B1
|
cytochrome P450 family 4 subfamily B member 1
|
|
chr5_-_168883333
Show fit
|
10.26 |
ENST00000404867.7
|
SLIT3
|
slit guidance ligand 3
|
|
chr4_+_165378998
Show fit
|
9.90 |
ENST00000402744.9
|
CPE
|
carboxypeptidase E
|
|
chr11_+_111912725
Show fit
|
9.35 |
ENST00000304298.4
|
HSPB2
|
heat shock protein family B (small) member 2
|
|
chr11_-_111912871
Show fit
|
8.46 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B
|
|
chr5_-_169300782
Show fit
|
8.15 |
ENST00000332966.8
|
SLIT3
|
slit guidance ligand 3
|
|
chr3_-_73624840
Show fit
|
7.36 |
ENST00000308537.4
ENST00000263666.9
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr21_-_26843063
Show fit
|
7.23 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1
|
|
chr19_+_676385
Show fit
|
7.13 |
ENST00000166139.9
|
FSTL3
|
follistatin like 3
|
|
chr21_-_26843012
Show fit
|
7.01 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1
|
|
chr4_-_5019437
Show fit
|
6.09 |
ENST00000506508.1
ENST00000509419.1
ENST00000307746.9
|
CYTL1
|
cytokine like 1
|
|
chr8_+_19939246
Show fit
|
5.71 |
ENST00000650287.1
|
LPL
|
lipoprotein lipase
|
|
chr20_+_37521206
Show fit
|
5.65 |
ENST00000346199.3
ENST00000647955.1
ENST00000649451.1
ENST00000649697.1
ENST00000649309.1
|
NNAT
|
neuronatin
|
|
chr16_+_31472130
Show fit
|
5.65 |
ENST00000394863.8
ENST00000565360.5
ENST00000361773.7
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr1_-_39639626
Show fit
|
5.63 |
ENST00000372852.4
|
HEYL
|
hes related family bHLH transcription factor with YRPW motif like
|
|
chr8_-_17697654
Show fit
|
5.20 |
ENST00000297488.10
|
MTUS1
|
microtubule associated scaffold protein 1
|
|
chr21_-_26573211
Show fit
|
5.08 |
ENST00000299340.9
ENST00000652641.2
|
CYYR1
|
cysteine and tyrosine rich 1
|
|
chr5_+_72107453
Show fit
|
4.79 |
ENST00000296755.12
ENST00000511641.2
|
MAP1B
|
microtubule associated protein 1B
|
|
chr2_+_26692686
Show fit
|
4.57 |
ENST00000620977.1
ENST00000302909.4
|
KCNK3
|
potassium two pore domain channel subfamily K member 3
|
|
chr3_+_154121366
Show fit
|
4.23 |
ENST00000465093.6
ENST00000496710.5
ENST00000465817.1
|
ARHGEF26
|
Rho guanine nucleotide exchange factor 26
|
|
chr3_+_50269140
Show fit
|
4.21 |
ENST00000616701.5
ENST00000433753.4
ENST00000611067.4
|
SEMA3B
|
semaphorin 3B
|
|
chr11_+_118607598
Show fit
|
4.17 |
ENST00000600882.6
ENST00000356063.9
|
PHLDB1
|
pleckstrin homology like domain family B member 1
|
|
chr20_+_44714835
Show fit
|
4.02 |
ENST00000372868.6
|
CCN5
|
cellular communication network factor 5
|
|
chr11_-_12008584
Show fit
|
3.90 |
ENST00000534511.5
ENST00000683431.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3
|
|
chr17_+_7023042
Show fit
|
3.89 |
ENST00000293805.10
|
BCL6B
|
BCL6B transcription repressor
|
|
chr5_-_122077152
Show fit
|
3.71 |
ENST00000513319.5
ENST00000503759.5
|
LOX
|
lysyl oxidase
|
|
chr1_-_171652675
Show fit
|
3.65 |
ENST00000037502.11
|
MYOC
|
myocilin
|
|
chr3_+_170418856
Show fit
|
3.53 |
ENST00000064724.8
ENST00000486975.1
|
CLDN11
ENSG00000285218.1
|
claudin 11
novel protein
|
|
chr8_+_30387064
Show fit
|
3.52 |
ENST00000523115.5
ENST00000519647.5
|
RBPMS
|
RNA binding protein, mRNA processing factor
|
|
chr11_-_11621991
Show fit
|
3.52 |
ENST00000227756.5
|
GALNT18
|
polypeptide N-acetylgalactosaminyltransferase 18
|
|
chr11_-_111911759
Show fit
|
3.47 |
ENST00000650687.2
|
CRYAB
|
crystallin alpha B
|
|
chr20_+_44714853
Show fit
|
3.45 |
ENST00000372865.4
|
CCN5
|
cellular communication network factor 5
|
|
chr22_-_42720861
Show fit
|
3.40 |
ENST00000642412.2
|
A4GALT
|
alpha 1,4-galactosyltransferase (P blood group)
|
|
chr21_-_26845402
Show fit
|
3.39 |
ENST00000284984.8
ENST00000676955.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1
|
|
chr10_-_88952763
Show fit
|
3.31 |
ENST00000224784.10
|
ACTA2
|
actin alpha 2, smooth muscle
|
|
chr22_-_42720813
Show fit
|
3.28 |
ENST00000381278.4
|
A4GALT
|
alpha 1,4-galactosyltransferase (P blood group)
|
|
chrX_+_91434829
Show fit
|
3.27 |
ENST00000312600.4
|
PABPC5
|
poly(A) binding protein cytoplasmic 5
|
|
chr7_+_130492066
Show fit
|
3.26 |
ENST00000223215.10
ENST00000437945.6
|
MEST
|
mesoderm specific transcript
|
|
chr20_+_63696643
Show fit
|
3.23 |
ENST00000369996.3
|
TNFRSF6B
|
TNF receptor superfamily member 6b
|
|
chr17_-_35795592
Show fit
|
3.13 |
ENST00000615136.4
ENST00000605424.6
ENST00000612672.1
|
MMP28
|
matrix metallopeptidase 28
|
|
chr17_-_76537699
Show fit
|
3.02 |
ENST00000293230.10
|
CYGB
|
cytoglobin
|
|
chr17_+_62627628
Show fit
|
3.02 |
ENST00000303375.10
|
MRC2
|
mannose receptor C type 2
|
|
chr11_-_8593940
Show fit
|
2.99 |
ENST00000315204.5
ENST00000396672.5
ENST00000431279.6
ENST00000418597.5
|
STK33
|
serine/threonine kinase 33
|
|
chr17_+_42682470
Show fit
|
2.84 |
ENST00000264638.9
|
CNTNAP1
|
contactin associated protein 1
|
|
chr2_+_158968608
Show fit
|
2.80 |
ENST00000263635.8
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr19_-_40413364
Show fit
|
2.76 |
ENST00000291825.11
ENST00000324001.8
|
PRX
|
periaxin
|
|
chr14_+_64540734
Show fit
|
2.72 |
ENST00000247207.7
|
HSPA2
|
heat shock protein family A (Hsp70) member 2
|
|
chr5_-_142325001
Show fit
|
2.71 |
ENST00000344120.4
ENST00000434127.3
|
SPRY4
|
sprouty RTK signaling antagonist 4
|
|
chr5_-_44389407
Show fit
|
2.71 |
ENST00000264664.5
|
FGF10
|
fibroblast growth factor 10
|
|
chr10_+_80413817
Show fit
|
2.70 |
ENST00000372187.9
|
PRXL2A
|
peroxiredoxin like 2A
|
|
chr2_+_216633411
Show fit
|
2.68 |
ENST00000233809.9
|
IGFBP2
|
insulin like growth factor binding protein 2
|
|
chr22_-_19524400
Show fit
|
2.59 |
ENST00000618236.2
|
CLDN5
|
claudin 5
|
|
chrX_-_17861236
Show fit
|
2.52 |
ENST00000331511.5
ENST00000415486.7
ENST00000451717.6
ENST00000545871.1
|
RAI2
|
retinoic acid induced 2
|
|
chr1_-_205813177
Show fit
|
2.40 |
ENST00000367137.4
|
SLC41A1
|
solute carrier family 41 member 1
|
|
chr19_-_42253888
Show fit
|
2.39 |
ENST00000593944.5
|
ERF
|
ETS2 repressor factor
|
|
chr11_-_75668566
Show fit
|
2.37 |
ENST00000526740.3
|
MAP6
|
microtubule associated protein 6
|
|
chr19_-_35135011
Show fit
|
2.34 |
ENST00000310123.8
|
LGI4
|
leucine rich repeat LGI family member 4
|
|
chr16_-_21440719
Show fit
|
2.30 |
ENST00000521589.5
|
NPIPB3
|
nuclear pore complex interacting protein family member B3
|
|
chr21_-_26573145
Show fit
|
2.23 |
ENST00000400043.3
|
CYYR1
|
cysteine and tyrosine rich 1
|
|
chr8_+_74824526
Show fit
|
2.21 |
ENST00000649643.1
ENST00000260113.7
|
PI15
|
peptidase inhibitor 15
|
|
chr2_-_85663393
Show fit
|
2.19 |
ENST00000494165.1
|
SFTPB
|
surfactant protein B
|
|
chr1_-_182604379
Show fit
|
2.19 |
ENST00000367558.6
|
RGS16
|
regulator of G protein signaling 16
|
|
chr17_-_76537630
Show fit
|
2.15 |
ENST00000589342.1
|
CYGB
|
cytoglobin
|
|
chr19_-_35134861
Show fit
|
2.10 |
ENST00000591633.2
|
LGI4
|
leucine rich repeat LGI family member 4
|
|
chr16_+_28863946
Show fit
|
2.09 |
ENST00000395532.8
|
SH2B1
|
SH2B adaptor protein 1
|
|
chr9_+_106863121
Show fit
|
2.04 |
ENST00000472574.1
ENST00000277225.10
|
ZNF462
|
zinc finger protein 462
|
|
chr4_-_186596770
Show fit
|
1.99 |
ENST00000512772.5
|
FAT1
|
FAT atypical cadherin 1
|
|
chr9_-_122227525
Show fit
|
1.96 |
ENST00000373755.6
ENST00000373754.6
|
LHX6
|
LIM homeobox 6
|
|
chr22_+_31093358
Show fit
|
1.95 |
ENST00000404574.5
|
SMTN
|
smoothelin
|
|
chr5_+_53480619
Show fit
|
1.95 |
ENST00000396947.7
ENST00000256759.8
|
FST
|
follistatin
|
|
chr1_-_44843240
Show fit
|
1.89 |
ENST00000372192.4
|
PTCH2
|
patched 2
|
|
chr9_+_128411715
Show fit
|
1.89 |
ENST00000420034.5
ENST00000372842.5
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr10_-_93601228
Show fit
|
1.85 |
ENST00000371464.8
|
RBP4
|
retinol binding protein 4
|
|
chr11_-_64759967
Show fit
|
1.80 |
ENST00000377432.7
ENST00000164139.4
|
PYGM
|
glycogen phosphorylase, muscle associated
|
|
chr7_+_116672187
Show fit
|
1.75 |
ENST00000318493.11
ENST00000397752.8
|
MET
|
MET proto-oncogene, receptor tyrosine kinase
|
|
chr7_-_82443715
Show fit
|
1.71 |
ENST00000356253.9
ENST00000423588.1
|
CACNA2D1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1
|
|
chr6_-_29633171
Show fit
|
1.69 |
ENST00000377034.9
|
GABBR1
|
gamma-aminobutyric acid type B receptor subunit 1
|
|
chrX_-_17860707
Show fit
|
1.69 |
ENST00000360011.5
|
RAI2
|
retinoic acid induced 2
|
|
chr1_+_183186238
Show fit
|
1.68 |
ENST00000493293.5
ENST00000264144.5
|
LAMC2
|
laminin subunit gamma 2
|
|
chr1_-_183418364
Show fit
|
1.67 |
ENST00000287713.7
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr11_+_76782250
Show fit
|
1.65 |
ENST00000533752.1
ENST00000612930.1
|
TSKU
|
tsukushi, small leucine rich proteoglycan
|
|
chr6_-_29633056
Show fit
|
1.64 |
ENST00000377016.8
|
GABBR1
|
gamma-aminobutyric acid type B receptor subunit 1
|
|
chr6_+_36948257
Show fit
|
1.61 |
ENST00000647861.1
ENST00000611814.4
|
PI16
|
peptidase inhibitor 16
|
|
chr17_-_7234262
Show fit
|
1.61 |
ENST00000575756.5
ENST00000575458.5
|
DVL2
|
dishevelled segment polarity protein 2
|
|
chr10_-_95290992
Show fit
|
1.61 |
ENST00000329399.7
|
PDLIM1
|
PDZ and LIM domain 1
|
|
chr7_+_90403386
Show fit
|
1.61 |
ENST00000287916.8
ENST00000394604.5
ENST00000496677.6
ENST00000394605.2
ENST00000480135.1
|
CLDN12
ENSG00000273299.1
|
claudin 12
novel transcript
|
|
chr11_-_75668434
Show fit
|
1.58 |
ENST00000434603.2
|
MAP6
|
microtubule associated protein 6
|
|
chr6_+_43771960
Show fit
|
1.55 |
ENST00000230480.10
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr9_-_34397800
Show fit
|
1.52 |
ENST00000297623.7
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr11_+_118607579
Show fit
|
1.52 |
ENST00000530708.4
|
PHLDB1
|
pleckstrin homology like domain family B member 1
|
|
chr9_+_113349514
Show fit
|
1.49 |
ENST00000374183.5
|
BSPRY
|
B-box and SPRY domain containing
|
|
chr12_-_6688859
Show fit
|
1.48 |
ENST00000542351.5
ENST00000538829.5
|
ZNF384
|
zinc finger protein 384
|
|
chr1_-_85465147
Show fit
|
1.47 |
ENST00000284031.13
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr10_-_75401746
Show fit
|
1.46 |
ENST00000372524.5
|
ZNF503
|
zinc finger protein 503
|
|
chr3_-_9878765
Show fit
|
1.46 |
ENST00000430427.6
ENST00000383817.5
ENST00000679265.1
|
CIDEC
|
cell death inducing DFFA like effector c
|
|
chr3_+_137998735
Show fit
|
1.45 |
ENST00000343735.8
|
CLDN18
|
claudin 18
|
|
chr9_-_127877665
Show fit
|
1.42 |
ENST00000644144.2
|
AK1
|
adenylate kinase 1
|
|
chr11_-_12009082
Show fit
|
1.40 |
ENST00000396505.7
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3
|
|
chr10_+_134465
Show fit
|
1.40 |
ENST00000439456.5
ENST00000397962.8
ENST00000309776.8
ENST00000397959.7
|
ZMYND11
|
zinc finger MYND-type containing 11
|
|
chr11_-_12009134
Show fit
|
1.38 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3
|
|
chr3_+_159764002
Show fit
|
1.37 |
ENST00000460298.3
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough
|
|
chr4_-_185775271
Show fit
|
1.35 |
ENST00000430503.5
ENST00000319454.10
ENST00000450341.5
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_-_34815257
Show fit
|
1.34 |
ENST00000374789.8
ENST00000374788.8
ENST00000346874.9
ENST00000374794.8
ENST00000350537.9
ENST00000374790.8
ENST00000374776.6
ENST00000374773.6
ENST00000545260.5
ENST00000545693.5
ENST00000340077.9
|
PARD3
|
par-3 family cell polarity regulator
|
|
chr5_+_80960694
Show fit
|
1.34 |
ENST00000638442.1
|
RASGRF2
|
Ras protein specific guanine nucleotide releasing factor 2
|
|
chr7_-_108456378
Show fit
|
1.33 |
ENST00000613830.4
ENST00000413765.6
ENST00000379028.8
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr3_+_113897470
Show fit
|
1.33 |
ENST00000440446.2
ENST00000488680.1
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr7_-_108456321
Show fit
|
1.33 |
ENST00000379024.8
ENST00000351718.8
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr15_+_39581068
Show fit
|
1.29 |
ENST00000397591.2
ENST00000260356.6
|
THBS1
|
thrombospondin 1
|
|
chr22_+_37805218
Show fit
|
1.27 |
ENST00000340857.4
|
H1-0
|
H1.0 linker histone
|
|
chrX_+_51893533
Show fit
|
1.26 |
ENST00000375722.5
ENST00000326587.12
ENST00000375695.2
|
MAGED1
|
MAGE family member D1
|
|
chr17_-_7234462
Show fit
|
1.25 |
ENST00000005340.10
|
DVL2
|
dishevelled segment polarity protein 2
|
|
chr2_+_24793394
Show fit
|
1.23 |
ENST00000380834.7
ENST00000260662.2
|
CENPO
|
centromere protein O
|
|
chr17_+_7023062
Show fit
|
1.21 |
ENST00000576705.1
|
BCL6B
|
BCL6B transcription repressor
|
|
chr6_-_29628038
Show fit
|
1.20 |
ENST00000355973.7
ENST00000377012.8
|
GABBR1
|
gamma-aminobutyric acid type B receptor subunit 1
|
|
chr4_-_185775890
Show fit
|
1.15 |
ENST00000437304.6
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr7_-_151410024
Show fit
|
1.15 |
ENST00000628331.1
ENST00000334493.11
|
WDR86
|
WD repeat domain 86
|
|
chr8_+_42152946
Show fit
|
1.14 |
ENST00000518421.5
ENST00000174653.3
ENST00000396926.8
ENST00000521280.5
ENST00000522288.5
|
AP3M2
|
adaptor related protein complex 3 subunit mu 2
|
|
chr6_+_162727941
Show fit
|
1.13 |
ENST00000366888.6
|
PACRG
|
parkin coregulated
|
|
chr22_-_30387078
Show fit
|
1.12 |
ENST00000215798.10
|
RNF215
|
ring finger protein 215
|
|
chr2_+_136765508
Show fit
|
1.11 |
ENST00000409968.6
|
THSD7B
|
thrombospondin type 1 domain containing 7B
|
|
chr12_-_6689244
Show fit
|
1.08 |
ENST00000361959.7
ENST00000436774.6
ENST00000544482.1
|
ZNF384
|
zinc finger protein 384
|
|
chr5_+_72107234
Show fit
|
1.08 |
ENST00000512974.5
|
MAP1B
|
microtubule associated protein 1B
|
|
chr4_+_76949743
Show fit
|
1.07 |
ENST00000502584.5
ENST00000264893.11
ENST00000510641.5
|
SEPTIN11
|
septin 11
|
|
chr9_+_34990250
Show fit
|
1.06 |
ENST00000454002.6
ENST00000545841.5
|
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5
|
|
chr12_-_106084023
Show fit
|
1.06 |
ENST00000553094.1
ENST00000549704.1
|
NUAK1
|
NUAK family kinase 1
|
|
chr3_-_24495532
Show fit
|
1.02 |
ENST00000643772.1
ENST00000642307.1
ENST00000645139.1
|
THRB
|
thyroid hormone receptor beta
|
|
chr15_-_93073706
Show fit
|
1.02 |
ENST00000425933.6
|
RGMA
|
repulsive guidance molecule BMP co-receptor a
|
|
chr18_+_23949847
Show fit
|
1.01 |
ENST00000588004.1
|
LAMA3
|
laminin subunit alpha 3
|
|
chr4_+_76949807
Show fit
|
1.00 |
ENST00000505788.5
ENST00000510515.5
ENST00000504637.5
|
SEPTIN11
|
septin 11
|
|
chr3_+_138010143
Show fit
|
0.99 |
ENST00000183605.10
|
CLDN18
|
claudin 18
|
|
chr11_+_57761787
Show fit
|
0.98 |
ENST00000358694.10
ENST00000361332.8
ENST00000361391.10
ENST00000361796.9
ENST00000415361.6
ENST00000426142.6
ENST00000428599.6
ENST00000525902.5
ENST00000526357.5
ENST00000526772.5
ENST00000527467.5
ENST00000528232.5
ENST00000528621.5
ENST00000529526.5
ENST00000529873.5
ENST00000529986.5
ENST00000530094.5
ENST00000530748.5
ENST00000531014.5
ENST00000532245.5
ENST00000532463.5
ENST00000532649.5
ENST00000532787.5
ENST00000532844.5
ENST00000533667.5
ENST00000534579.5
ENST00000399050.10
ENST00000526938.5
ENST00000674015.1
ENST00000531007.2
ENST00000673826.1
|
CTNND1
|
catenin delta 1
|
|
chr17_+_7705193
Show fit
|
0.97 |
ENST00000226091.3
|
EFNB3
|
ephrin B3
|
|
chr22_-_45977154
Show fit
|
0.97 |
ENST00000339464.9
|
WNT7B
|
Wnt family member 7B
|
|
chrX_+_71144818
Show fit
|
0.96 |
ENST00000536169.5
ENST00000358741.4
ENST00000395855.6
ENST00000374051.7
|
NLGN3
|
neuroligin 3
|
|
chr1_-_43367956
Show fit
|
0.95 |
ENST00000372458.8
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr2_+_24793098
Show fit
|
0.95 |
ENST00000473706.5
|
CENPO
|
centromere protein O
|
|
chr5_-_90409720
Show fit
|
0.94 |
ENST00000522864.5
ENST00000283122.8
ENST00000522083.5
ENST00000522565.5
ENST00000522842.1
|
CETN3
|
centrin 3
|
|
chr10_-_79444216
Show fit
|
0.91 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger CCHC-type containing 24
|
|
chr9_+_34989641
Show fit
|
0.91 |
ENST00000453597.8
ENST00000312316.9
ENST00000458263.6
ENST00000537321.5
ENST00000682809.1
ENST00000684748.1
|
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5
|
|
chr10_+_24466487
Show fit
|
0.90 |
ENST00000396446.5
ENST00000396445.5
ENST00000376451.4
|
KIAA1217
|
KIAA1217
|
|
chr6_-_110179623
Show fit
|
0.90 |
ENST00000265601.7
ENST00000447287.5
ENST00000392589.6
ENST00000444391.5
|
WASF1
|
WASP family member 1
|
|
chr6_+_125219804
Show fit
|
0.88 |
ENST00000524679.1
|
TPD52L1
|
TPD52 like 1
|
|
chr3_+_4493471
Show fit
|
0.88 |
ENST00000544951.6
ENST00000650294.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1
|
|
chr4_-_89837106
Show fit
|
0.88 |
ENST00000394986.5
ENST00000394991.8
ENST00000506244.5
ENST00000394989.6
ENST00000673902.1
ENST00000674129.1
|
SNCA
|
synuclein alpha
|
|
chr17_-_31858952
Show fit
|
0.87 |
ENST00000378634.6
|
COPRS
|
coordinator of PRMT5 and differentiation stimulator
|
|
chr17_+_76001338
Show fit
|
0.86 |
ENST00000425876.6
|
CDK3
|
cyclin dependent kinase 3
|
|
chr17_+_7886178
Show fit
|
0.86 |
ENST00000672274.1
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr16_-_85112390
Show fit
|
0.85 |
ENST00000629253.1
ENST00000539556.6
|
CIBAR2
|
CBY1 interacting BAR domain containing 2
|
|
chr12_-_6689450
Show fit
|
0.84 |
ENST00000355772.8
ENST00000417772.7
ENST00000319770.7
ENST00000396801.7
|
ZNF384
|
zinc finger protein 384
|
|
chr17_+_48054675
Show fit
|
0.84 |
ENST00000582155.5
ENST00000583378.5
ENST00000536222.5
|
NFE2L1
|
nuclear factor, erythroid 2 like 1
|
|
chrX_+_54809060
Show fit
|
0.84 |
ENST00000396224.1
|
MAGED2
|
MAGE family member D2
|
|
chr22_-_27801712
Show fit
|
0.84 |
ENST00000302326.5
|
MN1
|
MN1 proto-oncogene, transcriptional regulator
|
|
chr9_-_113340248
Show fit
|
0.83 |
ENST00000341761.8
ENST00000374195.7
ENST00000374193.9
|
WDR31
|
WD repeat domain 31
|
|
chr9_-_122228845
Show fit
|
0.83 |
ENST00000394319.8
ENST00000340587.7
|
LHX6
|
LIM homeobox 6
|
|
chr5_-_149379286
Show fit
|
0.81 |
ENST00000261796.4
|
IL17B
|
interleukin 17B
|
|
chr17_+_7884783
Show fit
|
0.81 |
ENST00000380358.9
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr19_+_3314403
Show fit
|
0.80 |
ENST00000641145.1
|
NFIC
|
nuclear factor I C
|
|
chrX_-_139204275
Show fit
|
0.79 |
ENST00000441825.8
|
FGF13
|
fibroblast growth factor 13
|
|
chr9_-_33264559
Show fit
|
0.79 |
ENST00000473781.1
ENST00000379704.7
ENST00000488499.1
|
BAG1
|
BAG cochaperone 1
|
|
chr4_+_71187269
Show fit
|
0.78 |
ENST00000425175.5
ENST00000351898.10
|
SLC4A4
|
solute carrier family 4 member 4
|
|
chr16_+_86578543
Show fit
|
0.77 |
ENST00000320241.5
|
FOXL1
|
forkhead box L1
|
|
chr3_-_185824966
Show fit
|
0.74 |
ENST00000457616.6
ENST00000346192.7
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2
|
|
chr9_+_40223317
Show fit
|
0.73 |
ENST00000611747.1
ENST00000377601.3
ENST00000654136.1
ENST00000659103.1
|
ANKRD20A2P
ENSG00000240240.9
|
ankyrin repeat domain 20 family member A2, pseudogene
novel transcript
|
|
chr16_+_55324191
Show fit
|
0.72 |
ENST00000290552.8
|
IRX6
|
iroquois homeobox 6
|
|
chr11_+_45146631
Show fit
|
0.69 |
ENST00000534751.3
ENST00000683152.1
|
PRDM11
|
PR/SET domain 11
|
|
chr12_-_6689359
Show fit
|
0.69 |
ENST00000683879.1
|
ZNF384
|
zinc finger protein 384
|
|
chr11_-_19201976
Show fit
|
0.68 |
ENST00000647990.1
ENST00000649235.1
ENST00000265968.9
ENST00000649842.1
|
CSRP3
|
cysteine and glycine rich protein 3
|
|
chr11_-_120120880
Show fit
|
0.67 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29
|
|
chrX_-_139205006
Show fit
|
0.66 |
ENST00000436198.5
ENST00000626909.2
|
FGF13
|
fibroblast growth factor 13
|
|
chr6_-_110179702
Show fit
|
0.64 |
ENST00000392587.6
|
WASF1
|
WASP family member 1
|
|
chr3_-_185825029
Show fit
|
0.64 |
ENST00000382199.7
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2
|
|
chrX_-_52069172
Show fit
|
0.62 |
ENST00000486010.1
ENST00000497164.5
ENST00000360134.10
ENST00000485287.5
ENST00000335504.9
|
MAGED4B
|
MAGE family member D4B
|
|
chr16_-_981259
Show fit
|
0.60 |
ENST00000563837.1
ENST00000563863.5
ENST00000565069.5
ENST00000570014.5
|
CEROX1
LMF1
|
cytoplasmic endogenous regulator of oxidative phosphorylation 1
lipase maturation factor 1
|
|
chr1_-_43368039
Show fit
|
0.60 |
ENST00000413844.3
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr3_-_16512895
Show fit
|
0.58 |
ENST00000449415.5
ENST00000441460.5
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr10_+_134703
Show fit
|
0.57 |
ENST00000509513.6
|
ZMYND11
|
zinc finger MYND-type containing 11
|
|
chr4_-_185775432
Show fit
|
0.57 |
ENST00000457247.5
ENST00000435480.5
ENST00000425679.5
ENST00000457934.5
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr17_+_48055945
Show fit
|
0.56 |
ENST00000580037.1
|
NFE2L1
|
nuclear factor, erythroid 2 like 1
|
|
chr1_-_43367689
Show fit
|
0.56 |
ENST00000621943.4
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr2_+_68467544
Show fit
|
0.55 |
ENST00000303795.9
|
APLF
|
aprataxin and PNKP like factor
|
|
chr2_-_132670194
Show fit
|
0.54 |
ENST00000397463.3
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr9_+_132410183
Show fit
|
0.53 |
ENST00000393216.3
ENST00000343036.6
|
CFAP77
|
cilia and flagella associated protein 77
|
|
chr12_+_4269771
Show fit
|
0.52 |
ENST00000676411.1
|
CCND2
|
cyclin D2
|
|
chr14_-_106211453
Show fit
|
0.52 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20
|
|
chr3_+_111071773
Show fit
|
0.52 |
ENST00000485303.6
|
NECTIN3
|
nectin cell adhesion molecule 3
|
|
chr9_+_33265013
Show fit
|
0.52 |
ENST00000223500.9
|
CHMP5
|
charged multivesicular body protein 5
|
|
chr9_+_33264848
Show fit
|
0.51 |
ENST00000419016.6
|
CHMP5
|
charged multivesicular body protein 5
|
|
chr7_+_90403506
Show fit
|
0.51 |
ENST00000427904.1
|
CLDN12
|
claudin 12
|
|
chr19_-_12957198
Show fit
|
0.50 |
ENST00000316939.3
|
GADD45GIP1
|
GADD45G interacting protein 1
|
|
chr13_-_30306837
Show fit
|
0.49 |
ENST00000414289.5
|
KATNAL1
|
katanin catalytic subunit A1 like 1
|
|
chr9_-_113056670
Show fit
|
0.49 |
ENST00000553380.1
ENST00000374227.8
|
ZFP37
|
ZFP37 zinc finger protein
|
|
chr1_+_6613722
Show fit
|
0.48 |
ENST00000377648.5
|
PHF13
|
PHD finger protein 13
|
|
chr8_-_41665261
Show fit
|
0.48 |
ENST00000522231.5
ENST00000314214.12
ENST00000348036.8
ENST00000522543.5
|
ANK1
|
ankyrin 1
|
|
chr1_+_32209074
Show fit
|
0.47 |
ENST00000409358.2
|
DCDC2B
|
doublecortin domain containing 2B
|
|
chr4_+_61202142
Show fit
|
0.47 |
ENST00000514591.5
|
ADGRL3
|
adhesion G protein-coupled receptor L3
|
|
chr17_+_75093287
Show fit
|
0.46 |
ENST00000538213.6
ENST00000584118.1
|
SLC16A5
|
solute carrier family 16 member 5
|