Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for ZKSCAN3
Z-value: 0.48
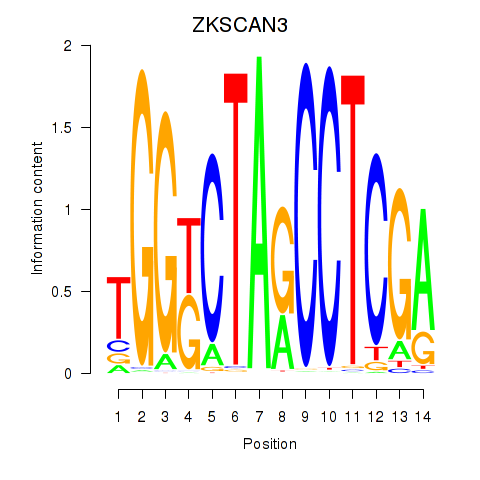
Transcription factors associated with ZKSCAN3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN3
|
ENSG00000189298.14 | zinc finger with KRAB and SCAN domains 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZKSCAN3 | hg38_v1_chr6_+_28349907_28349983 | -0.10 | 5.8e-01 | Click! |
Activity profile of ZKSCAN3 motif
Sorted Z-values of ZKSCAN3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_77017042 | 2.69 |
ENST00000359645.7
ENST00000397875.7 ENST00000397869.7 ENST00000578193.5 ENST00000578873.5 ENST00000397866.8 ENST00000528160.1 ENST00000527041.1 ENST00000526111.5 ENST00000397865.9 ENST00000382582.7 |
MBP
|
myelin basic protein |
| chr1_+_65792889 | 1.43 |
ENST00000341517.9
|
PDE4B
|
phosphodiesterase 4B |
| chr6_+_42929430 | 1.13 |
ENST00000372836.5
|
CNPY3
|
canopy FGF signaling regulator 3 |
| chr14_-_106130061 | 1.12 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr14_-_106675544 | 1.11 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr1_+_156153568 | 0.91 |
ENST00000368284.5
ENST00000368286.6 ENST00000368285.8 ENST00000438830.5 |
SEMA4A
|
semaphorin 4A |
| chr14_-_106593319 | 0.82 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr11_-_111449981 | 0.82 |
ENST00000531398.1
|
POU2AF1
|
POU class 2 homeobox associating factor 1 |
| chr7_-_151210488 | 0.78 |
ENST00000644661.2
|
H2BE1
|
H2B.E variant histone 1 |
| chr5_-_65722094 | 0.77 |
ENST00000381007.9
|
SGTB
|
small glutamine rich tetratricopeptide repeat containing beta |
| chr16_-_695946 | 0.67 |
ENST00000562563.1
|
FBXL16
|
F-box and leucine rich repeat protein 16 |
| chr1_-_19484635 | 0.67 |
ENST00000433834.5
|
CAPZB
|
capping actin protein of muscle Z-line subunit beta |
| chr1_+_226062704 | 0.58 |
ENST00000366814.3
ENST00000366815.10 ENST00000655399.1 ENST00000667897.1 |
H3-3A
|
H3.3 histone A |
| chr5_+_176810498 | 0.58 |
ENST00000509580.2
|
UNC5A
|
unc-5 netrin receptor A |
| chr2_+_190880809 | 0.56 |
ENST00000320717.8
|
GLS
|
glutaminase |
| chr15_-_72231583 | 0.54 |
ENST00000566809.1
ENST00000567087.5 ENST00000569050.1 ENST00000568883.5 |
PKM
|
pyruvate kinase M1/2 |
| chr1_+_113930371 | 0.47 |
ENST00000514621.5
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr5_+_65722190 | 0.45 |
ENST00000380985.10
ENST00000502464.5 |
NLN
|
neurolysin |
| chr14_-_106389858 | 0.45 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr15_-_72231113 | 0.44 |
ENST00000565154.6
ENST00000565184.6 ENST00000335181.10 |
PKM
|
pyruvate kinase M1/2 |
| chr14_+_64113084 | 0.41 |
ENST00000673797.1
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chr6_+_116529013 | 0.41 |
ENST00000628083.1
ENST00000368597.6 ENST00000452373.5 ENST00000405399.5 |
CALHM4
|
calcium homeostasis modulator family member 4 |
| chr6_+_32968557 | 0.40 |
ENST00000374825.9
|
BRD2
|
bromodomain containing 2 |
| chr5_+_176810552 | 0.39 |
ENST00000329542.9
|
UNC5A
|
unc-5 netrin receptor A |
| chr17_+_38752731 | 0.38 |
ENST00000619426.5
ENST00000610434.4 |
PSMB3
|
proteasome 20S subunit beta 3 |
| chrX_-_154547546 | 0.37 |
ENST00000440967.5
ENST00000369620.6 ENST00000393564.6 |
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr3_-_122793772 | 0.37 |
ENST00000306103.3
|
HSPBAP1
|
HSPB1 associated protein 1 |
| chr19_+_39296399 | 0.37 |
ENST00000333625.3
|
IFNL1
|
interferon lambda 1 |
| chr19_+_48755512 | 0.37 |
ENST00000593756.6
|
FGF21
|
fibroblast growth factor 21 |
| chr2_-_61017414 | 0.37 |
ENST00000421319.5
|
PUS10
|
pseudouridine synthase 10 |
| chr14_+_64113054 | 0.36 |
ENST00000673869.1
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chr1_-_19485468 | 0.35 |
ENST00000375142.5
|
CAPZB
|
capping actin protein of muscle Z-line subunit beta |
| chr12_-_3873382 | 0.34 |
ENST00000228820.9
ENST00000450737.2 ENST00000447133.7 |
PARP11
|
poly(ADP-ribose) polymerase family member 11 |
| chr21_+_38296556 | 0.34 |
ENST00000398927.1
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr2_+_190880834 | 0.34 |
ENST00000338435.8
|
GLS
|
glutaminase |
| chr1_-_19485502 | 0.33 |
ENST00000264203.7
ENST00000375144.6 ENST00000674432.1 ENST00000264202.8 |
CAPZB
|
capping actin protein of muscle Z-line subunit beta |
| chr16_+_2429427 | 0.33 |
ENST00000397066.9
|
CCNF
|
cyclin F |
| chr12_-_3873346 | 0.32 |
ENST00000427057.6
|
PARP11
|
poly(ADP-ribose) polymerase family member 11 |
| chr16_-_28495519 | 0.31 |
ENST00000569430.7
|
CLN3
|
CLN3 lysosomal/endosomal transmembrane protein, battenin |
| chr5_+_140700437 | 0.30 |
ENST00000274712.8
|
ZMAT2
|
zinc finger matrin-type 2 |
| chr5_-_180808742 | 0.29 |
ENST00000502678.1
|
MGAT1
|
alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase |
| chr1_+_113930728 | 0.27 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr17_-_30292042 | 0.27 |
ENST00000580709.1
|
BLMH
|
bleomycin hydrolase |
| chr6_+_12749419 | 0.25 |
ENST00000406205.7
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chrX_+_154547606 | 0.23 |
ENST00000594239.6
ENST00000615874.4 ENST00000619941.4 ENST00000617207.4 ENST00000611176.4 |
IKBKG
|
inhibitor of nuclear factor kappa B kinase regulatory subunit gamma |
| chr12_+_71754834 | 0.23 |
ENST00000261263.5
|
RAB21
|
RAB21, member RAS oncogene family |
| chr1_-_28088564 | 0.19 |
ENST00000373863.3
ENST00000373871.8 ENST00000540618.5 ENST00000436342.6 |
EYA3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr16_-_782054 | 0.19 |
ENST00000543963.5
|
MSLNL
|
mesothelin like |
| chrX_+_154547654 | 0.15 |
ENST00000615186.4
|
IKBKG
|
inhibitor of nuclear factor kappa B kinase regulatory subunit gamma |
| chr19_-_4182527 | 0.14 |
ENST00000601571.1
ENST00000601488.5 ENST00000305232.10 ENST00000337491.7 |
SIRT6
|
sirtuin 6 |
| chr19_-_2328573 | 0.13 |
ENST00000587502.2
ENST00000252622.15 ENST00000585409.2 |
LSM7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr21_+_38296909 | 0.13 |
ENST00000419868.1
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr20_-_59042748 | 0.11 |
ENST00000355937.9
ENST00000371033.9 |
PRELID3B
|
PRELI domain containing 3B |
| chrX_+_86148441 | 0.10 |
ENST00000373125.9
ENST00000373131.5 |
DACH2
|
dachshund family transcription factor 2 |
| chr12_-_113135710 | 0.08 |
ENST00000446861.7
|
RASAL1
|
RAS protein activator like 1 |
| chr1_-_99005003 | 0.07 |
ENST00000672681.1
ENST00000370188.7 |
PLPPR5
|
phospholipid phosphatase related 5 |
| chr22_-_49918404 | 0.06 |
ENST00000330817.11
|
ALG12
|
ALG12 alpha-1,6-mannosyltransferase |
| chr5_-_115262851 | 0.06 |
ENST00000379615.3
ENST00000419445.6 |
PGGT1B
|
protein geranylgeranyltransferase type I subunit beta |
| chr19_-_4182500 | 0.04 |
ENST00000597896.5
|
SIRT6
|
sirtuin 6 |
| chr1_+_3682306 | 0.04 |
ENST00000603362.5
ENST00000604479.5 |
TP73
|
tumor protein p73 |
| chr1_+_233327760 | 0.03 |
ENST00000366623.7
|
MAP3K21
|
mitogen-activated protein kinase kinase kinase 21 |
| chr16_-_1378989 | 0.02 |
ENST00000513783.1
|
UNKL
|
unk like zinc finger |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZKSCAN3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.2 | 1.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.5 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 1.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.3 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 1.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.8 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.4 | GO:0009051 | NADPH regeneration(GO:0006740) pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.4 | GO:1904640 | response to methionine(GO:1904640) |
| 0.1 | 0.2 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.0 | 0.7 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.4 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.3 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.9 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 3.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.2 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.2 | 2.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.4 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 1.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.8 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 2.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.4 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.1 | 0.2 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 1.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |