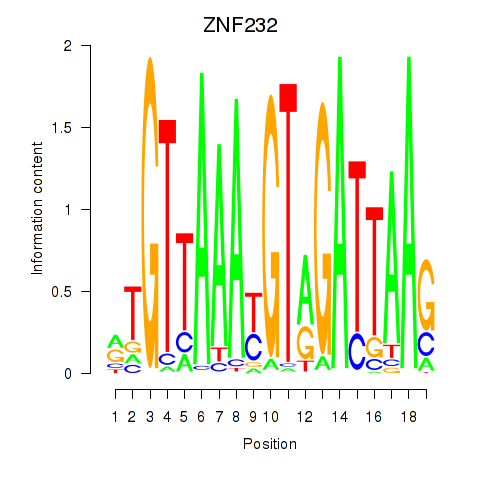
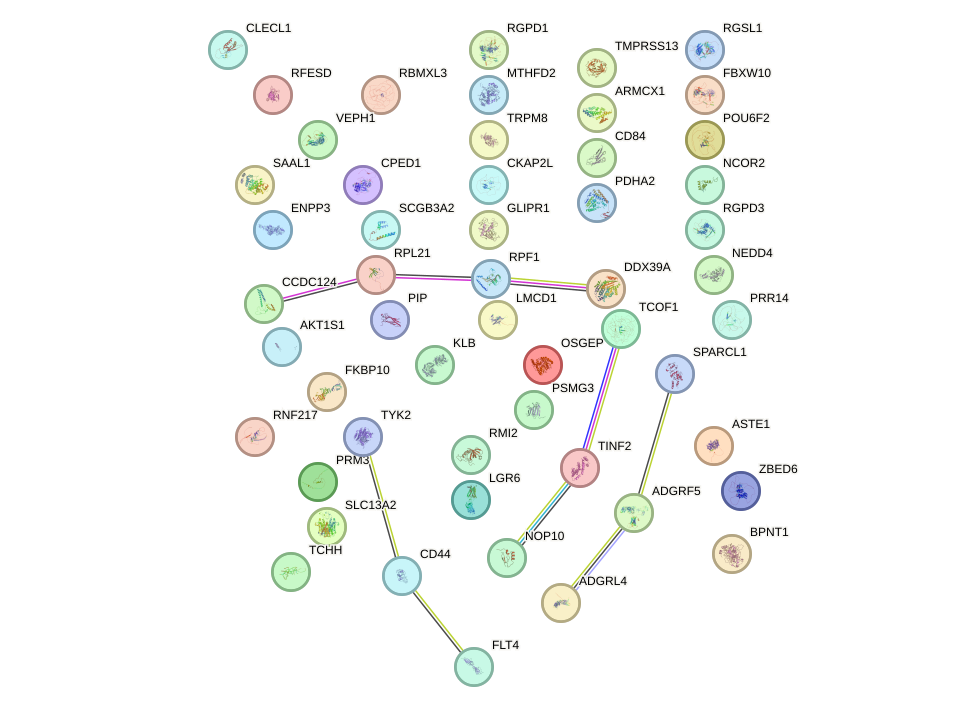
|
chr7_+_143132069
Show fit
|
2.58 |
ENST00000291009.4
|
PIP
|
prolactin induced protein
|
|
chr14_-_106675544
Show fit
|
2.58 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66
|
|
chr14_-_106269133
Show fit
|
2.40 |
ENST00000390609.3
|
IGHV3-23
|
immunoglobulin heavy variable 3-23
|
|
chr14_-_106360320
Show fit
|
2.17 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33
|
|
chr14_+_22547495
Show fit
|
2.02 |
ENST00000611116.2
|
TRAC
|
T cell receptor alpha constant
|
|
chr5_+_147878703
Show fit
|
1.76 |
ENST00000296694.5
|
SCGB3A2
|
secretoglobin family 3A member 2
|
|
chrY_+_12904860
Show fit
|
1.69 |
ENST00000336079.8
|
DDX3Y
|
DEAD-box helicase 3 Y-linked
|
|
chr5_-_180649579
Show fit
|
1.63 |
ENST00000261937.11
ENST00000502649.5
|
FLT4
|
fms related receptor tyrosine kinase 4
|
|
chrY_+_12904102
Show fit
|
1.61 |
ENST00000360160.8
ENST00000454054.5
|
DDX3Y
|
DEAD-box helicase 3 Y-linked
|
|
chr1_-_79006773
Show fit
|
1.54 |
ENST00000671209.1
|
ADGRL4
|
adhesion G protein-coupled receptor L4
|
|
chr5_-_180649613
Show fit
|
1.23 |
ENST00000393347.7
ENST00000619105.4
|
FLT4
|
fms related receptor tyrosine kinase 4
|
|
chr1_-_79006680
Show fit
|
1.23 |
ENST00000370742.4
ENST00000656841.1
|
ADGRL4
|
adhesion G protein-coupled receptor L4
|
|
chr3_+_8501807
Show fit
|
1.17 |
ENST00000426878.2
ENST00000397386.7
ENST00000415597.5
ENST00000157600.8
|
LMCD1
|
LIM and cysteine rich domains 1
|
|
chr6_-_46954922
Show fit
|
1.13 |
ENST00000265417.7
|
ADGRF5
|
adhesion G protein-coupled receptor F5
|
|
chr17_+_41819201
Show fit
|
0.92 |
ENST00000455106.1
|
FKBP10
|
FKBP prolyl isomerase 10
|
|
chr7_+_120989030
Show fit
|
0.88 |
ENST00000428526.5
|
CPED1
|
cadherin like and PC-esterase domain containing 1
|
|
chr1_+_203795614
Show fit
|
0.80 |
ENST00000367210.3
ENST00000432282.5
ENST00000453771.5
ENST00000367214.5
ENST00000639812.1
ENST00000367212.7
ENST00000332127.8
ENST00000550078.2
|
ZC3H11A
ZBED6
|
zinc finger CCCH-type containing 11A
zinc finger BED-type containing 6
|
|
chr12_-_9733292
Show fit
|
0.76 |
ENST00000621400.4
ENST00000327839.3
|
CLECL1
|
C-type lectin like 1
|
|
chr16_+_30651039
Show fit
|
0.75 |
ENST00000542965.2
|
PRR14
|
proline rich 14
|
|
chr4_-_87529460
Show fit
|
0.73 |
ENST00000418378.5
|
SPARCL1
|
SPARC like 1
|
|
chr2_+_74198605
Show fit
|
0.70 |
ENST00000409804.5
ENST00000678340.1
ENST00000679055.1
ENST00000394053.7
ENST00000409601.1
|
MTHFD2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase
|
|
chr16_+_30650932
Show fit
|
0.68 |
ENST00000300835.9
|
PRR14
|
proline rich 14
|
|
chr16_+_30650728
Show fit
|
0.63 |
ENST00000568754.5
|
PRR14
|
proline rich 14
|
|
chr6_-_49713564
Show fit
|
0.61 |
ENST00000616725.4
ENST00000618917.4
|
CRISP2
|
cysteine rich secretory protein 2
|
|
chr15_-_34343112
Show fit
|
0.58 |
ENST00000557912.1
ENST00000328848.6
|
NOP10
|
NOP10 ribonucleoprotein
|
|
chr19_-_14419300
Show fit
|
0.49 |
ENST00000589675.5
|
DDX39A
|
DExD-box helicase 39A
|
|
chr12_+_75481204
Show fit
|
0.49 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis related 1
|
|
chr6_-_49713521
Show fit
|
0.49 |
ENST00000339139.5
|
CRISP2
|
cysteine rich secretory protein 2
|
|
chr11_-_117929380
Show fit
|
0.44 |
ENST00000528626.5
ENST00000445164.6
ENST00000430170.6
ENST00000524993.6
ENST00000526090.1
|
TMPRSS13
|
transmembrane serine protease 13
|
|
chr6_-_26056460
Show fit
|
0.43 |
ENST00000343677.4
|
H1-2
|
H1.2 linker histone, cluster member
|
|
chr16_+_30650764
Show fit
|
0.43 |
ENST00000569864.5
|
PRR14
|
proline rich 14
|
|
chr6_+_124983356
Show fit
|
0.33 |
ENST00000519799.5
ENST00000368414.6
ENST00000359704.2
|
RNF217
|
ring finger protein 217
|
|
chr13_+_27251569
Show fit
|
0.30 |
ENST00000272274.8
ENST00000319826.8
ENST00000326092.8
|
RPL21
|
ribosomal protein L21
|
|
chr19_-_10380558
Show fit
|
0.30 |
ENST00000524462.5
ENST00000525621.6
ENST00000531836.5
|
TYK2
|
tyrosine kinase 2
|
|
chr5_+_150357629
Show fit
|
0.28 |
ENST00000650162.1
ENST00000377797.7
ENST00000445265.6
ENST00000323668.11
ENST00000643257.2
ENST00000646961.1
ENST00000513538.2
ENST00000439160.6
ENST00000394269.7
ENST00000427724.7
ENST00000504761.6
ENST00000513346.5
ENST00000515516.1
|
TCOF1
|
treacle ribosome biogenesis factor 1
|
|
chr2_-_112764600
Show fit
|
0.27 |
ENST00000302450.11
|
CKAP2L
|
cytoskeleton associated protein 2 like
|
|
chr1_-_160579439
Show fit
|
0.24 |
ENST00000368054.8
ENST00000368048.7
ENST00000311224.8
ENST00000368051.3
ENST00000534968.5
|
CD84
|
CD84 molecule
|
|
chr16_-_11273610
Show fit
|
0.23 |
ENST00000327157.4
|
PRM3
|
protamine 3
|
|
chr3_-_131026558
Show fit
|
0.22 |
ENST00000504725.1
ENST00000509060.1
|
ASTE1
|
asteroid homolog 1
|
|
chr4_+_95840084
Show fit
|
0.21 |
ENST00000295266.6
|
PDHA2
|
pyruvate dehydrogenase E1 subunit alpha 2
|
|
chr16_+_11345429
Show fit
|
0.18 |
ENST00000576027.1
ENST00000312499.6
ENST00000648619.1
|
RMI2
|
RecQ mediated genome instability 2
|
|
chr11_+_35189964
Show fit
|
0.18 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr19_+_17933001
Show fit
|
0.17 |
ENST00000445755.7
|
CCDC124
|
coiled-coil domain containing 124
|
|
chr19_-_14419035
Show fit
|
0.17 |
ENST00000590239.5
ENST00000590696.1
ENST00000591275.5
ENST00000586993.5
|
DDX39A
|
DExD-box helicase 39A
|
|
chr3_-_157503339
Show fit
|
0.17 |
ENST00000392833.6
|
VEPH1
|
ventricular zone expressed PH domain containing 1
|
|
chr6_+_131637296
Show fit
|
0.15 |
ENST00000358229.6
ENST00000357639.8
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr3_-_157503375
Show fit
|
0.14 |
ENST00000362010.7
|
VEPH1
|
ventricular zone expressed PH domain containing 1
|
|
chrX_+_101550537
Show fit
|
0.11 |
ENST00000372829.8
|
ARMCX1
|
armadillo repeat containing X-linked 1
|
|
chr19_-_49877279
Show fit
|
0.11 |
ENST00000391832.7
ENST00000344175.10
|
AKT1S1
|
AKT1 substrate 1
|
|
chr19_-_10380454
Show fit
|
0.10 |
ENST00000530829.1
ENST00000529370.5
|
TYK2
|
tyrosine kinase 2
|
|
chr2_+_86907953
Show fit
|
0.09 |
ENST00000409776.6
|
RGPD1
|
RANBP2 like and GRIP domain containing 1
|
|
chr7_-_1569955
Show fit
|
0.09 |
ENST00000288607.3
ENST00000404674.7
|
PSMG3
|
proteasome assembly chaperone 3
|
|
chr19_+_36111151
Show fit
|
0.07 |
ENST00000633214.1
ENST00000585332.3
|
OVOL3
|
ovo like zinc finger 3
|
|
chr1_-_152115443
Show fit
|
0.07 |
ENST00000614923.1
|
TCHH
|
trichohyalin
|
|
chr17_+_28473635
Show fit
|
0.06 |
ENST00000314669.10
ENST00000545060.2
|
SLC13A2
|
solute carrier family 13 member 2
|
|
chrX_+_115189410
Show fit
|
0.06 |
ENST00000424776.5
|
RBMXL3
|
RBMX like 3
|
|
chr14_-_20454741
Show fit
|
0.05 |
ENST00000206542.9
ENST00000553640.3
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr5_+_95646854
Show fit
|
0.05 |
ENST00000311364.8
ENST00000458310.1
|
RFESD
|
Rieske Fe-S domain containing
|
|
chr1_+_182450132
Show fit
|
0.04 |
ENST00000294854.13
|
RGSL1
|
regulator of G protein signaling like 1
|
|
chr4_+_39406917
Show fit
|
0.04 |
ENST00000257408.5
|
KLB
|
klotho beta
|
|
chr14_-_24242600
Show fit
|
0.04 |
ENST00000646753.1
ENST00000558566.1
ENST00000267415.12
ENST00000559019.1
ENST00000399423.8
ENST00000626689.2
|
TINF2
|
TERF1 interacting nuclear factor 2
|
|
chr2_+_233917371
Show fit
|
0.03 |
ENST00000324695.9
ENST00000433712.6
|
TRPM8
|
transient receptor potential cation channel subfamily M member 8
|
|
chr17_+_28473278
Show fit
|
0.03 |
ENST00000444914.7
|
SLC13A2
|
solute carrier family 13 member 2
|
|
chr1_+_202203721
Show fit
|
0.03 |
ENST00000255432.11
|
LGR6
|
leucine rich repeat containing G protein-coupled receptor 6
|
|
chr12_+_55549602
Show fit
|
0.02 |
ENST00000641569.1
ENST00000641851.1
|
OR6C4
|
olfactory receptor family 6 subfamily C member 4
|
|
chr17_+_18744026
Show fit
|
0.02 |
ENST00000308799.8
ENST00000395665.9
ENST00000301938.4
|
FBXW10
|
F-box and WD repeat domain containing 10
|
|
chr1_+_84479239
Show fit
|
0.01 |
ENST00000370656.5
ENST00000370654.6
|
RPF1
|
ribosome production factor 1 homolog
|
|
chr11_-_18106019
Show fit
|
0.00 |
ENST00000532452.5
ENST00000530180.1
ENST00000524803.6
ENST00000300013.8
ENST00000529318.5
|
SAAL1
|
serum amyloid A like 1
|