Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
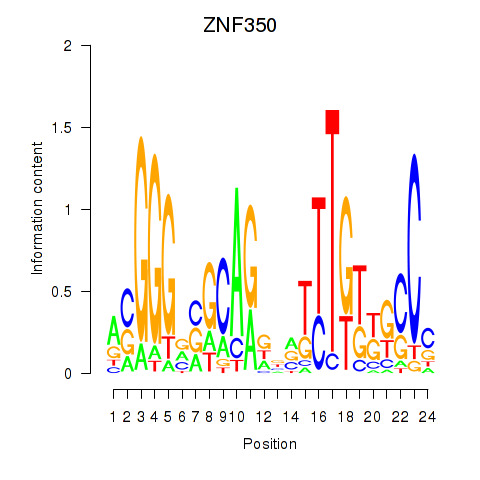
Results for ZNF350
Z-value: 0.74
Transcription factors associated with ZNF350
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF350
|
ENSG00000256683.7 | zinc finger protein 350 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF350 | hg38_v1_chr19_-_51986670_51986859 | 0.11 | 5.4e-01 | Click! |
Activity profile of ZNF350 motif
Sorted Z-values of ZNF350 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_167248638 | 2.77 |
ENST00000295237.10
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr1_+_228208054 | 2.00 |
ENST00000284548.16
ENST00000422127.5 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr10_-_15719885 | 1.69 |
ENST00000378076.4
|
ITGA8
|
integrin subunit alpha 8 |
| chr1_+_228208024 | 1.66 |
ENST00000570156.7
ENST00000680850.1 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr17_+_70075215 | 1.64 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr15_+_42402375 | 1.62 |
ENST00000397200.8
ENST00000569827.5 |
CAPN3
|
calpain 3 |
| chr19_-_17377334 | 1.57 |
ENST00000252590.9
ENST00000599426.1 |
PLVAP
|
plasmalemma vesicle associated protein |
| chr16_+_31214088 | 1.42 |
ENST00000613872.1
|
TRIM72
|
tripartite motif containing 72 |
| chr17_+_70075317 | 1.41 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr19_+_35041708 | 1.32 |
ENST00000672452.2
ENST00000673426.1 ENST00000600390.1 ENST00000597419.1 |
HPN
|
hepsin |
| chr16_+_31214111 | 1.27 |
ENST00000322122.8
|
TRIM72
|
tripartite motif containing 72 |
| chr5_-_16616972 | 1.26 |
ENST00000682564.1
ENST00000306320.10 ENST00000682229.1 |
RETREG1
|
reticulophagy regulator 1 |
| chr5_+_7396099 | 1.16 |
ENST00000338316.9
|
ADCY2
|
adenylate cyclase 2 |
| chr3_-_128493173 | 1.15 |
ENST00000498200.1
ENST00000341105.7 |
GATA2
|
GATA binding protein 2 |
| chr17_+_19127535 | 1.11 |
ENST00000577213.1
ENST00000344415.9 |
GRAPL
|
GRB2 related adaptor protein like |
| chr5_-_16617085 | 1.09 |
ENST00000684521.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr3_-_51968387 | 1.03 |
ENST00000490063.5
ENST00000468324.5 ENST00000497653.5 ENST00000484633.5 |
PCBP4
|
poly(rC) binding protein 4 |
| chr17_-_19046957 | 0.96 |
ENST00000284154.10
ENST00000573099.5 |
GRAP
|
GRB2 related adaptor protein |
| chr9_+_34992849 | 0.82 |
ENST00000443266.2
|
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr11_-_45665578 | 0.74 |
ENST00000308064.7
|
CHST1
|
carbohydrate sulfotransferase 1 |
| chr12_+_109131350 | 0.72 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr17_+_41812974 | 0.71 |
ENST00000321562.9
|
FKBP10
|
FKBP prolyl isomerase 10 |
| chr17_+_41812673 | 0.68 |
ENST00000585664.5
ENST00000585922.5 ENST00000429461.5 |
FKBP10
|
FKBP prolyl isomerase 10 |
| chr12_+_93378550 | 0.66 |
ENST00000550056.5
ENST00000549992.5 ENST00000548662.5 ENST00000547014.5 |
NUDT4
|
nudix hydrolase 4 |
| chr2_-_218659592 | 0.63 |
ENST00000411696.7
|
ZNF142
|
zinc finger protein 142 |
| chr20_+_45406560 | 0.60 |
ENST00000372717.5
ENST00000360981.8 |
DBNDD2
|
dysbindin domain containing 2 |
| chr17_-_41812586 | 0.59 |
ENST00000355468.7
ENST00000590496.1 |
P3H4
|
prolyl 3-hydroxylase family member 4 (inactive) |
| chr22_+_22343185 | 0.58 |
ENST00000427632.2
|
IGLV9-49
|
immunoglobulin lambda variable 9-49 |
| chr4_+_74933095 | 0.55 |
ENST00000513238.5
|
PARM1
|
prostate androgen-regulated mucin-like protein 1 |
| chr1_-_204685353 | 0.53 |
ENST00000367176.7
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr4_+_119212636 | 0.52 |
ENST00000274030.10
|
USP53
|
ubiquitin specific peptidase 53 |
| chr9_+_121567057 | 0.50 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr1_+_22643626 | 0.47 |
ENST00000374640.9
ENST00000374639.7 ENST00000374637.1 |
C1QC
|
complement C1q C chain |
| chr5_+_98769273 | 0.47 |
ENST00000308234.11
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr11_+_64555924 | 0.46 |
ENST00000301891.9
|
SLC22A11
|
solute carrier family 22 member 11 |
| chr2_-_218659468 | 0.44 |
ENST00000450560.1
ENST00000449707.5 ENST00000440934.2 |
ZNF142
|
zinc finger protein 142 |
| chr3_-_14178569 | 0.42 |
ENST00000285021.12
|
XPC
|
XPC complex subunit, DNA damage recognition and repair factor |
| chr12_+_93378625 | 0.42 |
ENST00000546925.1
|
NUDT4
|
nudix hydrolase 4 |
| chr12_+_6927946 | 0.42 |
ENST00000396684.3
|
ATN1
|
atrophin 1 |
| chr9_-_89436476 | 0.41 |
ENST00000420681.6
|
SEMA4D
|
semaphorin 4D |
| chr11_+_64555956 | 0.41 |
ENST00000377581.7
|
SLC22A11
|
solute carrier family 22 member 11 |
| chr19_-_48740573 | 0.41 |
ENST00000222145.9
|
RASIP1
|
Ras interacting protein 1 |
| chr16_-_46831134 | 0.38 |
ENST00000394806.6
ENST00000285697.9 |
C16orf87
|
chromosome 16 open reading frame 87 |
| chr12_-_113135710 | 0.36 |
ENST00000446861.7
|
RASAL1
|
RAS protein activator like 1 |
| chr1_+_156150008 | 0.35 |
ENST00000355014.6
|
SEMA4A
|
semaphorin 4A |
| chr17_-_8190154 | 0.34 |
ENST00000389017.6
|
BORCS6
|
BLOC-1 related complex subunit 6 |
| chr1_-_50960230 | 0.34 |
ENST00000396153.7
|
FAF1
|
Fas associated factor 1 |
| chr11_-_61920267 | 0.34 |
ENST00000531922.2
ENST00000301773.9 |
RAB3IL1
|
RAB3A interacting protein like 1 |
| chr15_+_90902218 | 0.32 |
ENST00000558290.5
ENST00000558853.5 ENST00000559999.5 |
MAN2A2
|
mannosidase alpha class 2A member 2 |
| chr11_-_119346695 | 0.31 |
ENST00000619721.6
|
MFRP
|
membrane frizzled-related protein |
| chr11_-_64744317 | 0.31 |
ENST00000419843.1
ENST00000394430.5 |
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr17_-_76453142 | 0.31 |
ENST00000319380.12
|
UBE2O
|
ubiquitin conjugating enzyme E2 O |
| chr15_-_42737054 | 0.30 |
ENST00000563260.1
ENST00000356231.4 |
CDAN1
|
codanin 1 |
| chr19_+_5681140 | 0.29 |
ENST00000579559.1
ENST00000577222.5 |
HSD11B1L
RPL36
|
hydroxysteroid 11-beta dehydrogenase 1 like ribosomal protein L36 |
| chr11_-_112164080 | 0.28 |
ENST00000528832.1
ENST00000280357.12 |
IL18
|
interleukin 18 |
| chr9_-_89436439 | 0.27 |
ENST00000418828.1
|
SEMA4D
|
semaphorin 4D |
| chr6_+_37005630 | 0.27 |
ENST00000274963.13
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr21_+_34668986 | 0.26 |
ENST00000349499.3
|
CLIC6
|
chloride intracellular channel 6 |
| chr14_+_96204679 | 0.25 |
ENST00000542454.2
ENST00000539359.1 ENST00000554311.2 ENST00000553811.1 |
BDKRB2
ENSG00000258691.1
|
bradykinin receptor B2 novel protein |
| chr11_-_112164056 | 0.24 |
ENST00000524595.5
|
IL18
|
interleukin 18 |
| chr9_+_129665603 | 0.24 |
ENST00000372469.6
|
PRRX2
|
paired related homeobox 2 |
| chr11_-_68903796 | 0.24 |
ENST00000362034.7
|
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr11_+_68903849 | 0.23 |
ENST00000675615.1
ENST00000255078.8 |
IGHMBP2
|
immunoglobulin mu DNA binding protein 2 |
| chr3_-_14178615 | 0.23 |
ENST00000511155.1
|
XPC
|
XPC complex subunit, DNA damage recognition and repair factor |
| chr4_-_25862979 | 0.23 |
ENST00000399878.8
|
SEL1L3
|
SEL1L family member 3 |
| chr22_+_21665994 | 0.23 |
ENST00000680393.1
ENST00000679534.1 ENST00000679827.1 ENST00000681956.1 ENST00000681338.1 ENST00000680061.1 ENST00000679540.1 ENST00000679795.1 ENST00000335025.12 ENST00000398831.8 ENST00000679477.1 ENST00000626352.2 ENST00000458567.5 ENST00000680094.1 ENST00000680109.1 ENST00000406385.1 ENST00000680860.1 |
PPIL2
|
peptidylprolyl isomerase like 2 |
| chr17_-_44014946 | 0.23 |
ENST00000587529.1
ENST00000206380.8 |
TMEM101
|
transmembrane protein 101 |
| chr11_+_8911154 | 0.21 |
ENST00000309357.8
ENST00000529876.5 ENST00000309377.9 ENST00000525005.5 ENST00000524577.5 ENST00000534506.5 |
AKIP1
|
A-kinase interacting protein 1 |
| chr12_+_8032692 | 0.21 |
ENST00000162391.8
|
FOXJ2
|
forkhead box J2 |
| chr11_+_706196 | 0.21 |
ENST00000534755.5
ENST00000650127.1 |
EPS8L2
|
EPS8 like 2 |
| chr8_-_42843201 | 0.21 |
ENST00000529779.1
ENST00000345117.2 ENST00000254250.7 |
THAP1
|
THAP domain containing 1 |
| chr17_-_81923532 | 0.21 |
ENST00000392366.7
|
MAFG
|
MAF bZIP transcription factor G |
| chr5_+_17604177 | 0.20 |
ENST00000510838.2
|
TAF11L11
|
TATA-box binding protein associated factor 11 like 11 |
| chr21_-_28992815 | 0.20 |
ENST00000361371.10
|
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr1_+_109249530 | 0.20 |
ENST00000271332.4
|
CELSR2
|
cadherin EGF LAG seven-pass G-type receptor 2 |
| chr16_+_67248966 | 0.20 |
ENST00000299798.16
|
SLC9A5
|
solute carrier family 9 member A5 |
| chr3_-_129183874 | 0.19 |
ENST00000422453.7
ENST00000451728.6 ENST00000446936.6 ENST00000502976.5 ENST00000500450.6 ENST00000441626.6 ENST00000504813.5 ENST00000512338.1 |
CNBP
|
CCHC-type zinc finger nucleic acid binding protein |
| chr11_+_118956289 | 0.19 |
ENST00000264031.3
|
UPK2
|
uroplakin 2 |
| chr21_-_28992947 | 0.18 |
ENST00000389194.7
ENST00000389195.7 ENST00000614971.4 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr10_+_67884646 | 0.18 |
ENST00000212015.11
|
SIRT1
|
sirtuin 1 |
| chr11_+_208815 | 0.17 |
ENST00000530889.1
ENST00000626818.1 |
RIC8A
|
RIC8 guanine nucleotide exchange factor A |
| chr18_-_5892104 | 0.17 |
ENST00000383490.2
|
TMEM200C
|
transmembrane protein 200C |
| chr15_-_58933668 | 0.17 |
ENST00000380516.7
|
SLTM
|
SAFB like transcription modulator |
| chr3_+_37861926 | 0.17 |
ENST00000443503.6
|
CTDSPL
|
CTD small phosphatase like |
| chr6_+_32969305 | 0.16 |
ENST00000456339.5
|
BRD2
|
bromodomain containing 2 |
| chr11_+_8911106 | 0.16 |
ENST00000299576.9
|
AKIP1
|
A-kinase interacting protein 1 |
| chr17_+_2337480 | 0.16 |
ENST00000268989.8
ENST00000426855.6 |
SGSM2
|
small G protein signaling modulator 2 |
| chr2_+_232633551 | 0.16 |
ENST00000264059.8
|
EFHD1
|
EF-hand domain family member D1 |
| chr6_+_32969345 | 0.15 |
ENST00000678250.1
|
BRD2
|
bromodomain containing 2 |
| chr1_-_155244645 | 0.15 |
ENST00000327247.9
|
GBA
|
glucosylceramidase beta |
| chr11_-_11621991 | 0.15 |
ENST00000227756.5
|
GALNT18
|
polypeptide N-acetylgalactosaminyltransferase 18 |
| chr10_+_123154768 | 0.15 |
ENST00000407911.2
|
BUB3
|
BUB3 mitotic checkpoint protein |
| chr19_-_5680488 | 0.15 |
ENST00000587589.1
ENST00000309324.9 |
MICOS13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chr11_+_706595 | 0.15 |
ENST00000531348.5
ENST00000530636.5 |
EPS8L2
|
EPS8 like 2 |
| chr19_+_2249317 | 0.14 |
ENST00000221496.5
|
AMH
|
anti-Mullerian hormone |
| chr6_-_56843153 | 0.14 |
ENST00000361203.7
ENST00000523817.1 |
DST
|
dystonin |
| chr10_-_12042771 | 0.13 |
ENST00000357604.10
|
UPF2
|
UPF2 regulator of nonsense mediated mRNA decay |
| chr17_+_2337622 | 0.13 |
ENST00000574563.5
|
SGSM2
|
small G protein signaling modulator 2 |
| chr10_-_92243246 | 0.12 |
ENST00000412050.8
ENST00000614585.4 |
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr10_-_119178791 | 0.12 |
ENST00000298510.4
|
PRDX3
|
peroxiredoxin 3 |
| chr11_-_68903777 | 0.12 |
ENST00000567045.5
ENST00000450904.6 |
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr17_+_50547089 | 0.11 |
ENST00000619622.4
ENST00000356488.8 ENST00000006658.11 |
SPATA20
|
spermatogenesis associated 20 |
| chr12_+_55931447 | 0.11 |
ENST00000549368.5
|
DGKA
|
diacylglycerol kinase alpha |
| chr16_+_69566314 | 0.11 |
ENST00000565301.2
|
NFAT5
|
nuclear factor of activated T cells 5 |
| chr19_-_55157725 | 0.11 |
ENST00000344887.10
ENST00000665070.1 |
TNNI3
|
troponin I3, cardiac type |
| chr16_+_78202 | 0.11 |
ENST00000356432.8
ENST00000219431.4 |
MPG
|
N-methylpurine DNA glycosylase |
| chr11_+_706222 | 0.11 |
ENST00000318562.13
ENST00000533500.5 |
EPS8L2
|
EPS8 like 2 |
| chr11_+_8911280 | 0.11 |
ENST00000530281.5
ENST00000396648.6 ENST00000534147.5 ENST00000529942.1 |
AKIP1
|
A-kinase interacting protein 1 |
| chr10_+_49614037 | 0.10 |
ENST00000395562.2
ENST00000337653.7 |
CHAT
|
choline O-acetyltransferase |
| chr1_+_63322558 | 0.10 |
ENST00000371116.4
|
FOXD3
|
forkhead box D3 |
| chr2_+_234952046 | 0.10 |
ENST00000416021.5
|
SH3BP4
|
SH3 domain binding protein 4 |
| chr6_+_32969165 | 0.10 |
ENST00000496118.2
ENST00000449085.4 |
BRD2
|
bromodomain containing 2 |
| chr15_-_58933559 | 0.10 |
ENST00000558486.5
ENST00000560682.5 ENST00000249736.11 ENST00000559880.5 ENST00000628684.2 |
SLTM
|
SAFB like transcription modulator |
| chr17_-_41440983 | 0.09 |
ENST00000246646.4
|
KRT38
|
keratin 38 |
| chr1_-_155244684 | 0.09 |
ENST00000428024.3
|
GBA
|
glucosylceramidase beta |
| chr11_-_535514 | 0.09 |
ENST00000451590.5
ENST00000311189.8 ENST00000417302.6 |
HRAS
|
HRas proto-oncogene, GTPase |
| chr3_+_37861849 | 0.08 |
ENST00000273179.10
|
CTDSPL
|
CTD small phosphatase like |
| chr11_-_119346655 | 0.08 |
ENST00000360167.4
|
MFRP
|
membrane frizzled-related protein |
| chr3_-_119034810 | 0.08 |
ENST00000393775.7
|
IGSF11
|
immunoglobulin superfamily member 11 |
| chr11_-_63768762 | 0.08 |
ENST00000433688.2
|
C11orf95
|
chromosome 11 open reading frame 95 |
| chr1_-_45623967 | 0.07 |
ENST00000445048.2
ENST00000421127.6 ENST00000528266.6 |
CCDC17
|
coiled-coil domain containing 17 |
| chr16_+_75148478 | 0.07 |
ENST00000568079.5
ENST00000570010.6 ENST00000464850.5 ENST00000332307.4 ENST00000393430.6 |
ZFP1
|
ZFP1 zinc finger protein |
| chr10_+_7818497 | 0.07 |
ENST00000344293.6
|
TAF3
|
TATA-box binding protein associated factor 3 |
| chr2_+_237692290 | 0.07 |
ENST00000420665.5
ENST00000392000.4 |
LRRFIP1
|
LRR binding FLII interacting protein 1 |
| chr20_+_19758245 | 0.06 |
ENST00000255006.12
|
RIN2
|
Ras and Rab interactor 2 |
| chr17_-_6651557 | 0.05 |
ENST00000225728.8
ENST00000575197.1 |
MED31
|
mediator complex subunit 31 |
| chr11_-_414948 | 0.05 |
ENST00000530494.1
ENST00000528209.5 ENST00000528058.1 ENST00000431843.7 |
SIGIRR
|
single Ig and TIR domain containing |
| chr3_-_119034779 | 0.05 |
ENST00000489689.5
|
IGSF11
|
immunoglobulin superfamily member 11 |
| chr2_-_156342348 | 0.05 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr11_+_706117 | 0.05 |
ENST00000533256.5
ENST00000614442.4 |
EPS8L2
|
EPS8 like 2 |
| chr5_+_17610496 | 0.04 |
ENST00000503184.2
|
TAF11L12
|
TATA-box binding protein associated factor 11 like 12 |
| chr12_-_108826161 | 0.04 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr12_+_130953898 | 0.04 |
ENST00000261654.10
|
ADGRD1
|
adhesion G protein-coupled receptor D1 |
| chr2_+_119759875 | 0.04 |
ENST00000263708.7
|
PTPN4
|
protein tyrosine phosphatase non-receptor type 4 |
| chr2_-_70553440 | 0.03 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha |
| chr15_-_65185299 | 0.03 |
ENST00000300107.7
|
CLPX
|
caseinolytic mitochondrial matrix peptidase chaperone subunit X |
| chr12_+_55932028 | 0.02 |
ENST00000394147.5
ENST00000551156.5 ENST00000553783.5 ENST00000557080.5 ENST00000432422.7 ENST00000556001.5 |
DGKA
|
diacylglycerol kinase alpha |
| chr11_-_64879675 | 0.02 |
ENST00000359393.6
ENST00000433803.1 ENST00000411683.1 |
EHD1
|
EH domain containing 1 |
| chr10_+_93060796 | 0.02 |
ENST00000651965.1
|
CYP26C1
|
cytochrome P450 family 26 subfamily C member 1 |
| chr10_+_123154414 | 0.02 |
ENST00000368858.9
|
BUB3
|
BUB3 mitotic checkpoint protein |
| chr2_+_241702027 | 0.01 |
ENST00000313552.11
ENST00000406941.5 |
ING5
|
inhibitor of growth family member 5 |
| chr3_+_12287859 | 0.01 |
ENST00000309576.11
ENST00000397015.7 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr3_+_141228721 | 0.01 |
ENST00000505013.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chr22_+_39077264 | 0.00 |
ENST00000407997.4
|
APOBEC3G
|
apolipoprotein B mRNA editing enzyme catalytic subunit 3G |
| chr5_+_177087412 | 0.00 |
ENST00000513166.1
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr1_-_246931326 | 0.00 |
ENST00000366508.5
ENST00000326225.3 |
AHCTF1
|
AT-hook containing transcription factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF350
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.4 | 1.3 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.3 | 2.4 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.3 | 1.7 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.3 | 3.7 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.2 | 1.1 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.2 | 2.7 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 1.1 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 0.5 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.2 | 1.6 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.1 | 0.7 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.7 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.2 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 1.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.5 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.2 | GO:1990619 | negative regulation of prostaglandin biosynthetic process(GO:0031393) histone H3-K9 deacetylation(GO:1990619) |
| 0.1 | 0.5 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 | 1.6 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 3.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.7 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 2.9 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.2 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.6 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.0 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 0.7 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.5 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 3.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 2.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 2.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 2.7 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 3.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 1.3 | GO:0005901 | caveola(GO:0005901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.2 | 5.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 3.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.7 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.1 | 0.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.3 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.1 | 0.2 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.9 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 1.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 2.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 2.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 1.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 3.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 2.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 3.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |