Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
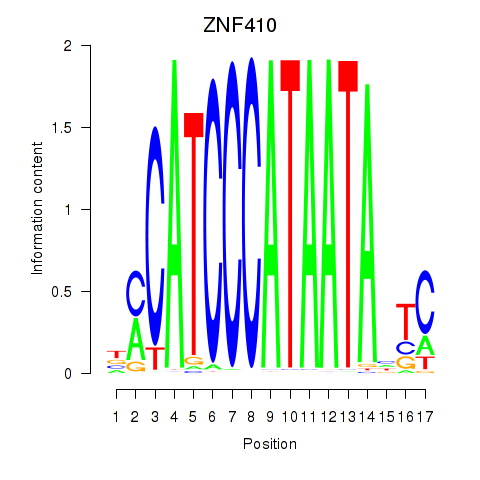
Results for ZNF410
Z-value: 0.42
Transcription factors associated with ZNF410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF410
|
ENSG00000119725.20 | zinc finger protein 410 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF410 | hg38_v1_chr14_+_73886617_73886766 | 0.10 | 6.0e-01 | Click! |
Activity profile of ZNF410 motif
Sorted Z-values of ZNF410 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_6607334 | 2.27 |
ENST00000645645.1
ENST00000357008.7 ENST00000544484.6 ENST00000544040.7 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr12_-_6607397 | 2.22 |
ENST00000645005.1
ENST00000646806.1 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr7_+_2656092 | 1.04 |
ENST00000429448.1
|
TTYH3
|
tweety family member 3 |
| chr12_+_121803991 | 1.04 |
ENST00000604567.6
ENST00000542440.5 |
SETD1B
|
SET domain containing 1B, histone lysine methyltransferase |
| chr11_-_57567260 | 0.85 |
ENST00000526659.1
ENST00000527022.1 |
UBE2L6
|
ubiquitin conjugating enzyme E2 L6 |
| chr19_+_47337060 | 0.83 |
ENST00000600626.1
|
C5AR2
|
complement component 5a receptor 2 |
| chr21_-_5973383 | 0.78 |
ENST00000464664.3
|
ENSG00000274559.3
|
novel histone H2B family protein |
| chr21_+_43565176 | 0.69 |
ENST00000599962.2
|
H2BS1
|
H2B.S histone 1 |
| chr1_+_206203541 | 0.62 |
ENST00000573034.8
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr22_-_38872206 | 0.62 |
ENST00000407418.8
ENST00000216083.6 |
CBX6
|
chromobox 6 |
| chr11_-_57567617 | 0.52 |
ENST00000287156.9
|
UBE2L6
|
ubiquitin conjugating enzyme E2 L6 |
| chr17_-_79797030 | 0.52 |
ENST00000269385.9
|
CBX8
|
chromobox 8 |
| chr10_+_27532521 | 0.51 |
ENST00000683924.1
|
RAB18
|
RAB18, member RAS oncogene family |
| chr4_-_83012924 | 0.34 |
ENST00000505397.1
|
LIN54
|
lin-54 DREAM MuvB core complex component |
| chr20_+_15196834 | 0.24 |
ENST00000402914.5
|
MACROD2
|
mono-ADP ribosylhydrolase 2 |
| chr8_-_81695045 | 0.22 |
ENST00000518568.3
|
SLC10A5
|
solute carrier family 10 member 5 |
| chr4_-_83012846 | 0.21 |
ENST00000510557.5
|
LIN54
|
lin-54 DREAM MuvB core complex component |
| chr14_-_21383960 | 0.19 |
ENST00000555943.1
|
SUPT16H
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr1_-_206204065 | 0.10 |
ENST00000367129.6
ENST00000468509.1 |
FAM72A
|
family with sequence similarity 72 member A |
| chr1_+_121184964 | 0.09 |
ENST00000367123.8
|
SRGAP2C
|
SLIT-ROBO Rho GTPase activating protein 2C |
| chr17_-_15563428 | 0.05 |
ENST00000584811.5
ENST00000419890.3 ENST00000518321.6 ENST00000438826.7 ENST00000225576.7 ENST00000428082.6 ENST00000522212.6 |
TVP23C
TVP23C-CDRT4
|
trans-golgi network vesicle protein 23 homolog C TVP23C-CDRT4 readthrough |
| chr1_-_121185539 | 0.02 |
ENST00000471903.6
|
FAM72B
|
family with sequence similarity 72 member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF410
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.5 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.3 | 0.8 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.1 | 1.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.7 | GO:0021816 | extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.5 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 1.0 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.6 | GO:0031519 | PcG protein complex(GO:0031519) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.1 | 1.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 4.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 1.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 1.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |