Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
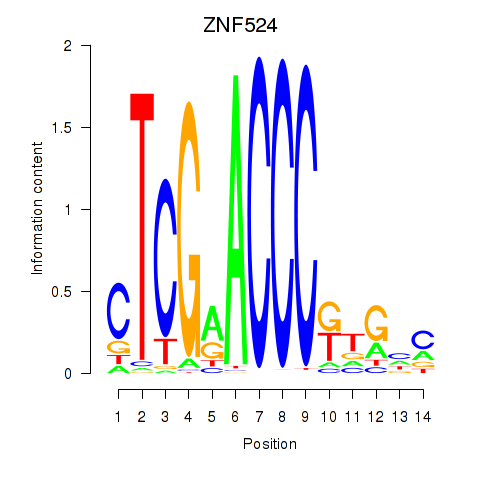
Results for ZNF524
Z-value: 1.39
Transcription factors associated with ZNF524
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF524
|
ENSG00000171443.7 | zinc finger protein 524 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF524 | hg38_v1_chr19_+_55600277_55600300 | 0.30 | 9.8e-02 | Click! |
Activity profile of ZNF524 motif
Sorted Z-values of ZNF524 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF524
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 1.3 | 11.3 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.9 | 2.7 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.7 | 2.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.6 | 2.4 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.5 | 1.4 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.5 | 1.4 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.5 | 2.3 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.4 | 1.3 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.4 | 1.2 | GO:0002339 | B cell selection(GO:0002339) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.4 | 1.6 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.4 | 1.9 | GO:1990426 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.3 | 1.7 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.3 | 1.0 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.3 | 1.0 | GO:0019483 | beta-alanine biosynthetic process(GO:0019483) |
| 0.3 | 2.5 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 | 2.5 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.3 | 0.8 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.3 | 0.3 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.2 | 2.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 0.9 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.2 | 0.2 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.2 | 0.9 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 0.9 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.2 | 0.9 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 0.6 | GO:0021658 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.2 | 1.0 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 0.8 | GO:1990918 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 | 0.6 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.2 | 0.6 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 1.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.2 | 0.2 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.2 | 2.3 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.2 | 0.6 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 0.6 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.2 | 2.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.2 | 0.5 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.2 | 0.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.2 | 0.7 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 | 1.5 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 1.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 0.5 | GO:1903123 | regulation of thioredoxin peroxidase activity(GO:1903123) negative regulation of thioredoxin peroxidase activity(GO:1903124) negative regulation of thioredoxin peroxidase activity by peptidyl-threonine phosphorylation(GO:1903125) Wnt signalosome assembly(GO:1904887) negative regulation of peroxidase activity(GO:2000469) |
| 0.2 | 1.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 1.0 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.2 | 1.9 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 1.0 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 1.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.9 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 3.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 3.2 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 1.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.6 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 3.0 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 0.4 | GO:0002465 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) |
| 0.1 | 1.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.4 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.5 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 0.4 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 2.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.4 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.1 | 0.5 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.8 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.5 | GO:0009631 | cold acclimation(GO:0009631) |
| 0.1 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.7 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.4 | GO:1904956 | neural crest cell fate commitment(GO:0014034) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 1.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.5 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 3.4 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 1.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 2.7 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.1 | 1.4 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 1.0 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.1 | 0.3 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.4 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.1 | 0.6 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.1 | 0.7 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 6.8 | GO:0007140 | male meiosis(GO:0007140) |
| 0.1 | 0.6 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 | 0.9 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 2.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.4 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 1.9 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.1 | 0.4 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.7 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.3 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 | 0.8 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.1 | 0.5 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.3 | GO:2001274 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 | 0.3 | GO:0002876 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.1 | 0.3 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 0.6 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 0.1 | 0.3 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 1.4 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.1 | 0.3 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.1 | 0.4 | GO:1903984 | rhythmic synaptic transmission(GO:0060024) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.3 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.1 | 0.5 | GO:0046778 | modification by virus of host mRNA processing(GO:0046778) |
| 0.1 | 1.0 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.6 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.3 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.1 | 1.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 1.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.3 | GO:1900100 | epithelial cell differentiation involved in salivary gland development(GO:0060690) epithelial cell maturation involved in salivary gland development(GO:0060691) regulation of plasma cell differentiation(GO:1900098) positive regulation of plasma cell differentiation(GO:1900100) positive regulation of lactation(GO:1903489) |
| 0.1 | 0.3 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 0.5 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 0.3 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.3 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 0.4 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.2 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.1 | 0.2 | GO:1904266 | positive regulation of Schwann cell migration(GO:1900149) regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.1 | 0.6 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 0.3 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.5 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.5 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 1.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.7 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.1 | 2.4 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.8 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.5 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.1 | 1.6 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.1 | 0.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.6 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.4 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.3 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 2.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 1.1 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.4 | GO:0003277 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) ureter morphogenesis(GO:0072197) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 3.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.2 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 1.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.3 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.1 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 2.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.6 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.5 | GO:0046086 | AMP catabolic process(GO:0006196) adenosine biosynthetic process(GO:0046086) |
| 0.1 | 1.6 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.1 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.1 | 0.3 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.5 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 0.6 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 0.6 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.1 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 1.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.7 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.2 | GO:0060482 | lobar bronchus development(GO:0060482) |
| 0.1 | 1.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 4.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.6 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.2 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 1.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.6 | GO:0032490 | detection of molecule of bacterial origin(GO:0032490) |
| 0.1 | 1.7 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 0.5 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 0.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.3 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 0.8 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.1 | 0.1 | GO:2000412 | regulation of thymocyte migration(GO:2000410) positive regulation of thymocyte migration(GO:2000412) |
| 0.1 | 2.7 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.3 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.1 | 1.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) |
| 0.1 | 1.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.2 | GO:0097212 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) |
| 0.1 | 0.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.3 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.7 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.1 | 0.6 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.1 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.4 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.7 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.2 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.2 | GO:2000830 | vacuolar phosphate transport(GO:0007037) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.1 | 0.3 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.1 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.3 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.8 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.3 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 1.0 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.0 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 1.0 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.2 | GO:0002864 | regulation of acute inflammatory response to antigenic stimulus(GO:0002864) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.0 | 0.1 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.8 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.2 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:1902263 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.3 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.9 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.6 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 1.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.5 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.2 | GO:0014062 | regulation of serotonin secretion(GO:0014062) |
| 0.0 | 0.1 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) RNA repair(GO:0042245) |
| 0.0 | 1.6 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.6 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.4 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 0.5 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.6 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.4 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.2 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 1.6 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.2 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 4.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.8 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.4 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.7 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.6 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.2 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 1.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.8 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.2 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.0 | 0.2 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.0 | 0.2 | GO:1901569 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.6 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.3 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.5 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.4 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 2.3 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.2 | GO:1902661 | positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 16.3 | GO:0007283 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.3 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.1 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.0 | 0.1 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.6 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.4 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.4 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.0 | 2.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 1.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.6 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.0 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.8 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.4 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 2.1 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.8 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.2 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 2.4 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 2.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 2.7 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.9 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.0 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.7 | GO:0070873 | regulation of glycogen metabolic process(GO:0070873) |
| 0.0 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.2 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 1.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.0 | GO:2001311 | lysobisphosphatidic acid metabolic process(GO:2001311) |
| 0.0 | 0.4 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.5 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:2000176 | pro-T cell differentiation(GO:0002572) regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.2 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.2 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.0 | GO:0070316 | regulation of G0 to G1 transition(GO:0070316) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 2.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.4 | 2.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.4 | 2.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.4 | 1.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 2.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.3 | 1.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 1.3 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.3 | 0.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 2.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 0.9 | GO:0036024 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.2 | 0.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.2 | 1.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 0.5 | GO:0032473 | cytoplasmic side of mitochondrial outer membrane(GO:0032473) caveola neck(GO:0099400) |
| 0.2 | 2.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 2.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 2.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.1 | 2.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.4 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.9 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.4 | GO:0043260 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.1 | 0.5 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 0.4 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 3.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 1.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.7 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.6 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.3 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.1 | 0.3 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 3.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.0 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 1.0 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 0.7 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.1 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 2.4 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 0.4 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 1.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 1.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 2.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.7 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 4.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 1.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 2.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 2.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 1.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 2.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.5 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 3.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 3.4 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 1.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0042825 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.0 | 0.4 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.8 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 2.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 2.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0071203 | F-actin capping protein complex(GO:0008290) WASH complex(GO:0071203) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 1.0 | 4.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.8 | 2.3 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.6 | 2.5 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.6 | 2.5 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.5 | 1.6 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.4 | 1.4 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.3 | 1.3 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.3 | 1.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.3 | 1.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.2 | 1.4 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 2.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 1.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 0.7 | GO:0047750 | cholestenol delta-isomerase activity(GO:0047750) |
| 0.2 | 0.9 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.2 | 0.7 | GO:0031696 | alpha2-adrenergic receptor activity(GO:0004938) alpha-2C adrenergic receptor binding(GO:0031696) |
| 0.2 | 0.9 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 0.8 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 0.6 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.2 | 0.6 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.2 | 0.6 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 0.5 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.2 | 0.5 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.2 | 0.5 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 1.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 0.5 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 0.5 | GO:0036479 | GTP-dependent protein kinase activity(GO:0034211) peroxidase inhibitor activity(GO:0036479) |
| 0.2 | 0.8 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 1.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 4.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 0.9 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.2 | 0.6 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 0.6 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 1.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.7 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 2.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.7 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 3.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.7 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.6 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.5 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 2.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 1.4 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.1 | 0.3 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.1 | 0.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.6 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 1.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 2.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.4 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.3 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.1 | 0.4 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.3 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 0.6 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.1 | 1.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.5 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 2.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 1.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.3 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 1.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 1.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 2.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 3.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.6 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.7 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.4 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.5 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.2 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.5 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.2 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 0.6 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 1.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.2 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 2.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.9 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.3 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.2 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.1 | 1.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.2 | GO:0052815 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.1 | 0.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.2 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 0.2 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.4 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 1.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 0.3 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 2.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.5 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.3 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 2.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.5 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 1.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 1.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 1.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 1.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.7 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 1.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 2.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.3 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 6.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 2.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 2.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 4.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 1.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.9 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.0 | 0.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 2.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 4.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.0 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 3.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 4.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 6.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 5.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 2.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 3.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 3.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 3.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 5.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 5.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.5 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 3.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 2.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 2.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 8.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 3.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.7 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.0 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 6.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 2.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.3 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |