Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
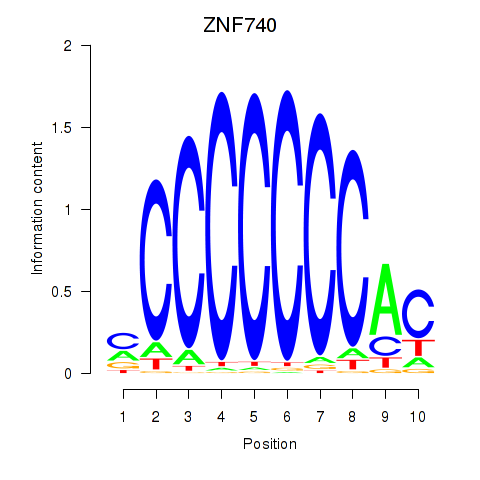
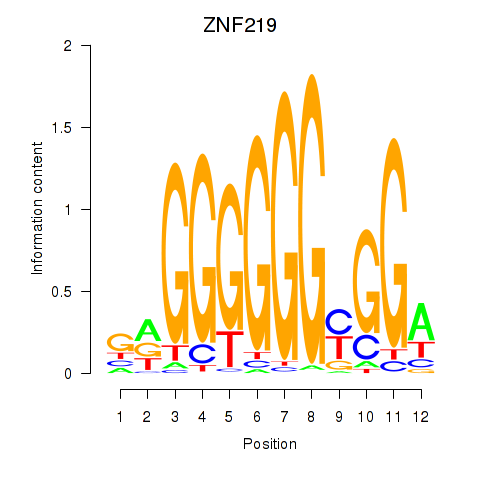
Results for ZNF740_ZNF219
Z-value: 1.60
Transcription factors associated with ZNF740_ZNF219
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF740
|
ENSG00000139651.11 | zinc finger protein 740 |
|
ZNF219
|
ENSG00000165804.16 | zinc finger protein 219 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF219 | hg38_v1_chr14_-_21098848_21099014 | -0.51 | 2.9e-03 | Click! |
| ZNF740 | hg38_v1_chr12_+_53180679_53180755 | 0.38 | 3.4e-02 | Click! |
Activity profile of ZNF740_ZNF219 motif
Sorted Z-values of ZNF740_ZNF219 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_29806519 | 5.83 |
ENST00000322945.11
ENST00000562337.5 ENST00000566906.6 ENST00000563402.1 ENST00000219782.10 |
MAZ
|
MYC associated zinc finger protein |
| chr7_+_21428184 | 5.04 |
ENST00000649633.1
|
SP4
|
Sp4 transcription factor |
| chr17_+_57256514 | 4.52 |
ENST00000284073.7
ENST00000674964.1 |
MSI2
|
musashi RNA binding protein 2 |
| chr7_+_21428023 | 4.32 |
ENST00000432066.2
ENST00000222584.8 |
SP4
|
Sp4 transcription factor |
| chr2_+_218399838 | 4.00 |
ENST00000273062.7
|
CTDSP1
|
CTD small phosphatase 1 |
| chr5_+_134525649 | 3.98 |
ENST00000282605.8
ENST00000681547.2 ENST00000361895.6 ENST00000402835.5 |
JADE2
|
jade family PHD finger 2 |
| chr16_+_30664334 | 3.71 |
ENST00000287468.5
|
FBRS
|
fibrosin |
| chr17_+_7884783 | 3.56 |
ENST00000380358.9
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr12_-_57846686 | 3.48 |
ENST00000548823.1
ENST00000398073.7 |
CTDSP2
|
CTD small phosphatase 2 |
| chr12_-_6606320 | 3.14 |
ENST00000642594.1
ENST00000644289.1 ENST00000645095.1 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr2_+_180980807 | 2.81 |
ENST00000602475.5
|
UBE2E3
|
ubiquitin conjugating enzyme E2 E3 |
| chr19_-_35742431 | 2.61 |
ENST00000592537.5
ENST00000246532.6 ENST00000588992.5 |
IGFLR1
|
IGF like family receptor 1 |
| chr6_-_32190170 | 2.59 |
ENST00000375050.6
|
PBX2
|
PBX homeobox 2 |
| chr16_+_29806078 | 2.57 |
ENST00000545521.5
|
MAZ
|
MYC associated zinc finger protein |
| chr2_+_218400039 | 2.55 |
ENST00000452977.5
|
CTDSP1
|
CTD small phosphatase 1 |
| chr11_-_67374168 | 2.54 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin like cytokine factor 1 |
| chr12_-_6606642 | 2.43 |
ENST00000545584.2
ENST00000545942.6 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr12_+_57522801 | 2.43 |
ENST00000355673.8
ENST00000546632.1 ENST00000549623.1 |
MBD6
|
methyl-CpG binding domain protein 6 |
| chr18_+_57435366 | 2.33 |
ENST00000491143.3
|
ONECUT2
|
one cut homeobox 2 |
| chr12_-_6606427 | 2.26 |
ENST00000642879.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr3_-_171460368 | 2.26 |
ENST00000436636.7
ENST00000465393.1 ENST00000341852.10 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr15_-_61229297 | 2.24 |
ENST00000335670.11
|
RORA
|
RAR related orphan receptor A |
| chr12_+_57755060 | 2.20 |
ENST00000266643.6
|
MARCHF9
|
membrane associated ring-CH-type finger 9 |
| chr5_+_138352674 | 2.18 |
ENST00000314358.10
|
KDM3B
|
lysine demethylase 3B |
| chr6_-_32192630 | 2.16 |
ENST00000375040.8
|
GPSM3
|
G protein signaling modulator 3 |
| chr19_-_39833615 | 2.14 |
ENST00000593685.5
ENST00000600611.5 |
DYRK1B
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr17_+_38705482 | 2.10 |
ENST00000620609.4
|
MLLT6
|
MLLT6, PHD finger containing |
| chr11_+_67119245 | 2.08 |
ENST00000529006.7
ENST00000398645.6 |
KDM2A
|
lysine demethylase 2A |
| chr1_-_33182030 | 2.05 |
ENST00000291416.10
|
TRIM62
|
tripartite motif containing 62 |
| chr16_+_30957960 | 2.04 |
ENST00000684162.1
|
SETD1A
|
SET domain containing 1A, histone lysine methyltransferase |
| chr12_+_53380639 | 2.04 |
ENST00000426431.2
|
SP1
|
Sp1 transcription factor |
| chr12_+_53985065 | 2.03 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr6_+_20403679 | 2.02 |
ENST00000535432.2
|
E2F3
|
E2F transcription factor 3 |
| chr17_+_57256727 | 2.01 |
ENST00000675656.1
|
MSI2
|
musashi RNA binding protein 2 |
| chr17_+_44557476 | 1.99 |
ENST00000315323.5
|
FZD2
|
frizzled class receptor 2 |
| chr16_+_30957714 | 1.96 |
ENST00000262519.14
ENST00000682768.1 |
SETD1A
|
SET domain containing 1A, histone lysine methyltransferase |
| chr1_+_26529745 | 1.93 |
ENST00000374168.7
ENST00000374166.8 |
RPS6KA1
|
ribosomal protein S6 kinase A1 |
| chr6_+_42782020 | 1.91 |
ENST00000314073.9
|
BICRAL
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr11_-_33869816 | 1.89 |
ENST00000395833.7
|
LMO2
|
LIM domain only 2 |
| chr3_-_171460063 | 1.89 |
ENST00000284483.12
ENST00000475336.5 ENST00000357327.9 ENST00000460047.5 ENST00000488470.5 ENST00000470834.5 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr6_-_32192845 | 1.89 |
ENST00000487761.5
|
GPSM3
|
G protein signaling modulator 3 |
| chr7_+_74658004 | 1.88 |
ENST00000443166.5
|
GTF2I
|
general transcription factor IIi |
| chr1_-_111204343 | 1.82 |
ENST00000369752.5
|
DENND2D
|
DENN domain containing 2D |
| chr16_+_81444799 | 1.81 |
ENST00000537098.8
|
CMIP
|
c-Maf inducing protein |
| chr1_+_26695993 | 1.81 |
ENST00000324856.13
ENST00000637465.1 |
ARID1A
|
AT-rich interaction domain 1A |
| chr12_+_57460127 | 1.81 |
ENST00000532291.5
ENST00000543426.5 ENST00000546141.5 |
GLI1
|
GLI family zinc finger 1 |
| chr17_+_67825664 | 1.79 |
ENST00000321892.8
|
BPTF
|
bromodomain PHD finger transcription factor |
| chr6_-_42451261 | 1.78 |
ENST00000372917.8
ENST00000340840.6 ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr12_-_6607397 | 1.78 |
ENST00000645005.1
ENST00000646806.1 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr4_+_153466324 | 1.77 |
ENST00000409663.7
ENST00000409959.8 |
TMEM131L
|
transmembrane 131 like |
| chr16_+_30698209 | 1.76 |
ENST00000411466.6
|
SRCAP
|
Snf2 related CREBBP activator protein |
| chr17_-_44219660 | 1.76 |
ENST00000436088.6
|
UBTF
|
upstream binding transcription factor |
| chr3_-_185824966 | 1.76 |
ENST00000457616.6
ENST00000346192.7 |
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr10_-_125160499 | 1.75 |
ENST00000494626.6
ENST00000337195.9 |
CTBP2
|
C-terminal binding protein 2 |
| chr12_-_6607334 | 1.75 |
ENST00000645645.1
ENST00000357008.7 ENST00000544484.6 ENST00000544040.7 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr1_-_150236150 | 1.73 |
ENST00000629042.2
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr14_-_36520222 | 1.71 |
ENST00000354822.7
|
NKX2-1
|
NK2 homeobox 1 |
| chr2_-_213150236 | 1.67 |
ENST00000442445.1
ENST00000342002.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr11_-_67373584 | 1.65 |
ENST00000543494.1
ENST00000312438.8 |
ENSG00000256514.1
CLCF1
|
novel protein cardiotrophin like cytokine factor 1 |
| chr8_-_56211257 | 1.65 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chr10_+_22321056 | 1.63 |
ENST00000376663.8
|
BMI1
|
BMI1 proto-oncogene, polycomb ring finger |
| chr12_+_57459782 | 1.62 |
ENST00000228682.7
|
GLI1
|
GLI family zinc finger 1 |
| chr1_-_52552994 | 1.61 |
ENST00000355809.4
ENST00000528642.5 ENST00000470626.1 ENST00000257177.9 ENST00000371544.7 |
TUT4
|
terminal uridylyl transferase 4 |
| chrX_-_143635081 | 1.61 |
ENST00000338017.8
|
SLITRK4
|
SLIT and NTRK like family member 4 |
| chr1_+_26696348 | 1.57 |
ENST00000457599.6
|
ARID1A
|
AT-rich interaction domain 1A |
| chr1_-_52553387 | 1.55 |
ENST00000484723.6
ENST00000524582.1 |
TUT4
|
terminal uridylyl transferase 4 |
| chr1_+_155002630 | 1.55 |
ENST00000535420.5
ENST00000417934.6 ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr1_-_150235943 | 1.54 |
ENST00000533654.5
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr15_-_56243829 | 1.54 |
ENST00000559447.8
ENST00000673997.1 |
RFX7
|
regulatory factor X7 |
| chr19_-_38617928 | 1.54 |
ENST00000396857.7
ENST00000586296.5 |
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr1_+_147541491 | 1.54 |
ENST00000683836.1
ENST00000234739.8 |
BCL9
|
BCL9 transcription coactivator |
| chr3_-_185825029 | 1.52 |
ENST00000382199.7
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr1_-_201023694 | 1.52 |
ENST00000332129.6
ENST00000422435.2 ENST00000461742.7 |
KIF21B
|
kinesin family member 21B |
| chr2_+_180981108 | 1.51 |
ENST00000602710.5
|
UBE2E3
|
ubiquitin conjugating enzyme E2 E3 |
| chr20_+_58891302 | 1.51 |
ENST00000371095.7
ENST00000265620.11 ENST00000354359.12 ENST00000371085.8 |
GNAS
|
GNAS complex locus |
| chr17_-_49362206 | 1.48 |
ENST00000430262.3
|
ZNF652
|
zinc finger protein 652 |
| chr1_-_145962581 | 1.47 |
ENST00000619813.1
|
ANKRD34A
|
ankyrin repeat domain 34A |
| chr7_+_50304693 | 1.47 |
ENST00000331340.8
ENST00000413698.5 ENST00000612658.4 ENST00000359197.9 ENST00000349824.8 ENST00000343574.9 ENST00000357364.8 ENST00000440768.6 ENST00000346667.8 ENST00000615491.4 |
IKZF1
|
IKAROS family zinc finger 1 |
| chr3_+_184315763 | 1.46 |
ENST00000456033.5
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma 1 |
| chr19_-_51646800 | 1.45 |
ENST00000599649.5
ENST00000429354.3 ENST00000360844.6 |
SIGLEC5
SIGLEC14
|
sialic acid binding Ig like lectin 5 sialic acid binding Ig like lectin 14 |
| chr12_+_50764509 | 1.45 |
ENST00000552487.1
|
ATF1
|
activating transcription factor 1 |
| chr5_+_56815534 | 1.44 |
ENST00000399503.4
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr17_+_56593685 | 1.43 |
ENST00000332822.6
|
NOG
|
noggin |
| chr2_+_111120906 | 1.43 |
ENST00000337565.9
ENST00000357757.6 ENST00000308659.12 ENST00000393256.8 ENST00000610735.4 ENST00000615946.4 ENST00000619294.4 ENST00000620862.4 ENST00000621302.4 ENST00000622509.4 ENST00000622612.4 |
BCL2L11
|
BCL2 like 11 |
| chr7_+_50304038 | 1.41 |
ENST00000642219.1
ENST00000645066.1 |
IKZF1
|
IKAROS family zinc finger 1 |
| chr1_+_43650466 | 1.41 |
ENST00000463151.5
|
KDM4A
|
lysine demethylase 4A |
| chr2_+_180981036 | 1.41 |
ENST00000602499.5
|
UBE2E3
|
ubiquitin conjugating enzyme E2 E3 |
| chr12_-_57251169 | 1.39 |
ENST00000554578.5
ENST00000546246.2 ENST00000332782.7 ENST00000553489.1 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr1_-_150235972 | 1.39 |
ENST00000534220.1
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr3_-_125375249 | 1.39 |
ENST00000484491.5
ENST00000492394.5 ENST00000471196.1 ENST00000468369.5 ENST00000485866.5 ENST00000360647.9 |
ZNF148
|
zinc finger protein 148 |
| chr6_-_151391539 | 1.39 |
ENST00000325144.5
|
ZBTB2
|
zinc finger and BTB domain containing 2 |
| chr17_-_44219728 | 1.39 |
ENST00000393606.7
|
UBTF
|
upstream binding transcription factor |
| chr5_-_132490750 | 1.38 |
ENST00000437654.6
ENST00000245414.9 ENST00000680139.1 ENST00000680352.1 ENST00000679440.1 ENST00000680903.1 |
IRF1
|
interferon regulatory factor 1 |
| chr13_-_40666600 | 1.36 |
ENST00000379561.6
|
FOXO1
|
forkhead box O1 |
| chr19_-_38617912 | 1.35 |
ENST00000591517.5
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr11_-_3840829 | 1.35 |
ENST00000396978.1
|
RHOG
|
ras homolog family member G |
| chr20_+_58891981 | 1.33 |
ENST00000488652.6
ENST00000476935.6 ENST00000492907.6 ENST00000603546.2 |
GNAS
|
GNAS complex locus |
| chr9_-_124507382 | 1.33 |
ENST00000373588.9
ENST00000620110.4 |
NR5A1
|
nuclear receptor subfamily 5 group A member 1 |
| chr19_-_4065732 | 1.33 |
ENST00000601588.1
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr3_+_107524357 | 1.32 |
ENST00000456817.5
ENST00000458458.5 |
BBX
|
BBX high mobility group box domain containing |
| chr9_-_20622479 | 1.31 |
ENST00000380338.9
|
MLLT3
|
MLLT3 super elongation complex subunit |
| chr6_-_41940537 | 1.30 |
ENST00000512426.5
|
CCND3
|
cyclin D3 |
| chr20_-_22584547 | 1.30 |
ENST00000419308.7
|
FOXA2
|
forkhead box A2 |
| chr18_-_76495191 | 1.29 |
ENST00000443185.7
|
ZNF516
|
zinc finger protein 516 |
| chr2_-_61471062 | 1.28 |
ENST00000398571.7
|
USP34
|
ubiquitin specific peptidase 34 |
| chr20_-_62065834 | 1.28 |
ENST00000252996.9
|
TAF4
|
TATA-box binding protein associated factor 4 |
| chr1_-_150236064 | 1.27 |
ENST00000532744.2
ENST00000369114.9 ENST00000369115.3 ENST00000583931.6 |
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr4_+_2793056 | 1.27 |
ENST00000503219.5
|
SH3BP2
|
SH3 domain binding protein 2 |
| chr6_+_32969305 | 1.27 |
ENST00000456339.5
|
BRD2
|
bromodomain containing 2 |
| chr6_-_142945028 | 1.26 |
ENST00000012134.7
|
HIVEP2
|
HIVEP zinc finger 2 |
| chrX_-_130110679 | 1.25 |
ENST00000335997.11
|
ELF4
|
E74 like ETS transcription factor 4 |
| chr17_+_57257002 | 1.24 |
ENST00000322684.7
ENST00000579590.5 |
MSI2
|
musashi RNA binding protein 2 |
| chr2_-_148021490 | 1.24 |
ENST00000416719.5
ENST00000264169.6 |
ORC4
|
origin recognition complex subunit 4 |
| chr3_+_184315347 | 1.24 |
ENST00000424196.5
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma 1 |
| chr3_-_18424533 | 1.24 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr12_-_121579638 | 1.24 |
ENST00000446152.6
|
KDM2B
|
lysine demethylase 2B |
| chr12_+_53380141 | 1.21 |
ENST00000551969.5
ENST00000327443.9 |
SP1
|
Sp1 transcription factor |
| chr17_+_14301069 | 1.19 |
ENST00000360954.3
|
HS3ST3B1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
| chr12_+_53985138 | 1.18 |
ENST00000303460.5
|
HOXC10
|
homeobox C10 |
| chr12_-_57129001 | 1.17 |
ENST00000556155.5
|
STAT6
|
signal transducer and activator of transcription 6 |
| chr20_+_32358979 | 1.17 |
ENST00000646985.1
ENST00000497249.6 |
ASXL1
|
ASXL transcriptional regulator 1 |
| chr6_+_18155329 | 1.16 |
ENST00000546309.6
|
KDM1B
|
lysine demethylase 1B |
| chr6_-_142945160 | 1.16 |
ENST00000367603.8
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr4_-_139177185 | 1.16 |
ENST00000394235.6
|
ELF2
|
E74 like ETS transcription factor 2 |
| chr6_-_31815244 | 1.15 |
ENST00000375654.5
|
HSPA1L
|
heat shock protein family A (Hsp70) member 1 like |
| chr15_-_41116211 | 1.15 |
ENST00000648947.1
|
INO80
|
INO80 complex ATPase subunit |
| chrX_+_123961696 | 1.15 |
ENST00000371145.8
ENST00000371157.7 ENST00000371144.7 |
STAG2
|
stromal antigen 2 |
| chr6_-_41940690 | 1.14 |
ENST00000505064.1
|
CCND3
|
cyclin D3 |
| chr15_+_40440889 | 1.14 |
ENST00000416165.6
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr19_-_38426195 | 1.14 |
ENST00000615439.5
ENST00000614135.4 ENST00000622174.4 ENST00000587753.5 ENST00000454404.6 ENST00000617966.4 ENST00000618320.4 ENST00000293062.13 ENST00000433821.6 ENST00000426920.6 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr7_-_105691637 | 1.14 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7 like 1 |
| chr1_+_178725277 | 1.12 |
ENST00000324778.5
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr3_+_184315131 | 1.12 |
ENST00000427845.5
ENST00000342981.8 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma 1 |
| chr20_-_4015389 | 1.12 |
ENST00000336095.10
|
RNF24
|
ring finger protein 24 |
| chr17_-_28950597 | 1.11 |
ENST00000583747.1
ENST00000584236.1 |
PHF12
|
PHD finger protein 12 |
| chr1_+_155063710 | 1.10 |
ENST00000359751.8
ENST00000368409.8 ENST00000427683.2 ENST00000505139.1 |
EFNA4
ENSG00000251246.1
|
ephrin A4 novel ephrin-A4 (EFNA4) and ephrin-A3 (EFNA3) protein |
| chr6_+_18155399 | 1.10 |
ENST00000650836.2
ENST00000449850.2 ENST00000297792.9 |
KDM1B
|
lysine demethylase 1B |
| chr2_+_69915100 | 1.10 |
ENST00000264444.7
|
MXD1
|
MAX dimerization protein 1 |
| chr20_-_20712626 | 1.10 |
ENST00000202677.12
|
RALGAPA2
|
Ral GTPase activating protein catalytic subunit alpha 2 |
| chr10_-_124093582 | 1.10 |
ENST00000462406.1
ENST00000435907.6 |
CHST15
|
carbohydrate sulfotransferase 15 |
| chr17_+_67825494 | 1.10 |
ENST00000306378.11
ENST00000544778.6 |
BPTF
|
bromodomain PHD finger transcription factor |
| chr1_-_153922742 | 1.09 |
ENST00000634408.1
ENST00000634791.1 |
GATAD2B
|
GATA zinc finger domain containing 2B |
| chr12_-_53207241 | 1.09 |
ENST00000267082.10
ENST00000549086.2 |
ITGB7
|
integrin subunit beta 7 |
| chr1_+_43650118 | 1.07 |
ENST00000372396.4
|
KDM4A
|
lysine demethylase 4A |
| chr6_+_32968557 | 1.06 |
ENST00000374825.9
|
BRD2
|
bromodomain containing 2 |
| chrX_-_130110479 | 1.06 |
ENST00000308167.10
|
ELF4
|
E74 like ETS transcription factor 4 |
| chr19_+_47608942 | 1.06 |
ENST00000594866.2
|
BICRA
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr5_+_50667405 | 1.06 |
ENST00000505554.5
|
PARP8
|
poly(ADP-ribose) polymerase family member 8 |
| chr1_+_178725227 | 1.05 |
ENST00000367635.8
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr12_+_55743283 | 1.05 |
ENST00000546799.1
|
GDF11
|
growth differentiation factor 11 |
| chr14_-_36519679 | 1.05 |
ENST00000498187.6
|
NKX2-1
|
NK2 homeobox 1 |
| chr7_-_155002911 | 1.05 |
ENST00000419436.1
ENST00000397192.5 |
PAXIP1
|
PAX interacting protein 1 |
| chr2_+_69915041 | 1.05 |
ENST00000540449.5
|
MXD1
|
MAX dimerization protein 1 |
| chr1_-_145962955 | 1.04 |
ENST00000619519.1
|
ANKRD34A
|
ankyrin repeat domain 34A |
| chr6_+_149317695 | 1.04 |
ENST00000637181.2
|
TAB2
|
TGF-beta activated kinase 1 (MAP3K7) binding protein 2 |
| chr17_-_62065248 | 1.03 |
ENST00000397786.7
|
MED13
|
mediator complex subunit 13 |
| chr5_+_50666612 | 1.02 |
ENST00000281631.10
|
PARP8
|
poly(ADP-ribose) polymerase family member 8 |
| chr12_-_53207271 | 1.02 |
ENST00000552972.5
ENST00000422257.7 |
ITGB7
|
integrin subunit beta 7 |
| chr19_-_41928449 | 1.02 |
ENST00000597630.3
|
ERFL
|
ETS repressor factor like |
| chr17_-_7251286 | 1.02 |
ENST00000576613.5
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr15_-_41116132 | 1.02 |
ENST00000616814.4
ENST00000401393.7 |
INO80
|
INO80 complex ATPase subunit |
| chr6_+_32969165 | 1.01 |
ENST00000496118.2
ENST00000449085.4 |
BRD2
|
bromodomain containing 2 |
| chr2_-_173964180 | 1.01 |
ENST00000418194.7
|
SP3
|
Sp3 transcription factor |
| chr12_-_57738740 | 1.00 |
ENST00000547588.6
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr1_-_150235995 | 1.00 |
ENST00000436748.6
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr19_-_10231271 | 1.00 |
ENST00000592342.5
ENST00000588952.5 |
DNMT1
|
DNA methyltransferase 1 |
| chr4_+_56907876 | 1.00 |
ENST00000640168.2
ENST00000309042.12 |
REST
|
RE1 silencing transcription factor |
| chr7_+_7968787 | 1.00 |
ENST00000223145.10
|
GLCCI1
|
glucocorticoid induced 1 |
| chr19_-_39834127 | 1.00 |
ENST00000601972.1
ENST00000430012.6 ENST00000323039.10 ENST00000348817.7 |
DYRK1B
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr12_-_9760893 | 0.99 |
ENST00000228434.7
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr20_-_63627049 | 0.99 |
ENST00000370077.2
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr6_+_15248855 | 0.99 |
ENST00000397311.4
|
JARID2
|
jumonji and AT-rich interaction domain containing 2 |
| chr14_-_24146314 | 0.98 |
ENST00000559056.5
|
PSME2
|
proteasome activator subunit 2 |
| chr12_-_57111338 | 0.98 |
ENST00000538913.6
ENST00000537215.6 ENST00000300134.8 ENST00000454075.7 ENST00000640254.2 ENST00000553275.1 ENST00000553533.2 |
STAT6
|
signal transducer and activator of transcription 6 |
| chr19_-_42132465 | 0.97 |
ENST00000529067.5
ENST00000529952.5 ENST00000342301.8 ENST00000389341.9 |
POU2F2
|
POU class 2 homeobox 2 |
| chr16_+_50742059 | 0.96 |
ENST00000311559.13
ENST00000564326.5 ENST00000566206.5 ENST00000427738.8 |
CYLD
|
CYLD lysine 63 deubiquitinase |
| chr1_-_211579064 | 0.96 |
ENST00000367001.5
|
SLC30A1
|
solute carrier family 30 member 1 |
| chr14_+_104800573 | 0.96 |
ENST00000555360.1
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr20_+_50731571 | 0.95 |
ENST00000371610.7
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr6_-_44127365 | 0.95 |
ENST00000372014.5
|
MRPL14
|
mitochondrial ribosomal protein L14 |
| chr14_+_22883220 | 0.95 |
ENST00000536884.1
ENST00000267396.9 |
REM2
|
RRAD and GEM like GTPase 2 |
| chr17_+_40121955 | 0.95 |
ENST00000398532.9
|
MSL1
|
MSL complex subunit 1 |
| chr17_+_45161070 | 0.95 |
ENST00000593138.6
ENST00000586681.6 |
HEXIM2
|
HEXIM P-TEFb complex subunit 2 |
| chrX_+_129980965 | 0.94 |
ENST00000607874.1
|
BCORL1
|
BCL6 corepressor like 1 |
| chr19_-_42132391 | 0.94 |
ENST00000528894.8
ENST00000560804.6 ENST00000560558.5 ENST00000560398.5 ENST00000526816.6 ENST00000625670.2 |
POU2F2
|
POU class 2 homeobox 2 |
| chr6_+_32969345 | 0.94 |
ENST00000678250.1
|
BRD2
|
bromodomain containing 2 |
| chr4_+_56908094 | 0.93 |
ENST00000622863.4
ENST00000514063.2 ENST00000638187.2 ENST00000616975.5 ENST00000640343.2 |
REST
|
RE1 silencing transcription factor |
| chr11_-_3840942 | 0.93 |
ENST00000351018.5
|
RHOG
|
ras homolog family member G |
| chr8_+_122781621 | 0.93 |
ENST00000314393.6
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr17_-_68291116 | 0.93 |
ENST00000327268.8
ENST00000580666.6 |
SLC16A6
|
solute carrier family 16 member 6 |
| chrX_-_40096190 | 0.93 |
ENST00000679513.1
|
BCOR
|
BCL6 corepressor |
| chr10_+_102776237 | 0.92 |
ENST00000369889.5
|
WBP1L
|
WW domain binding protein 1 like |
| chr15_-_61229155 | 0.92 |
ENST00000558904.2
|
RORA
|
RAR related orphan receptor A |
| chr15_+_100602519 | 0.92 |
ENST00000332783.12
ENST00000558747.5 ENST00000343276.4 |
ASB7
|
ankyrin repeat and SOCS box containing 7 |
| chr12_-_116277677 | 0.92 |
ENST00000281928.9
|
MED13L
|
mediator complex subunit 13L |
| chr17_-_7251691 | 0.91 |
ENST00000574322.6
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chrX_+_123961304 | 0.91 |
ENST00000371160.5
ENST00000435103.5 |
STAG2
|
stromal antigen 2 |
| chr15_-_70096604 | 0.91 |
ENST00000559048.5
ENST00000560939.5 ENST00000440567.7 ENST00000557907.5 ENST00000558379.5 ENST00000559929.5 |
TLE3
|
TLE family member 3, transcriptional corepressor |
| chr12_+_45729899 | 0.91 |
ENST00000422737.6
|
ARID2
|
AT-rich interaction domain 2 |
| chr6_-_41941507 | 0.90 |
ENST00000372987.8
|
CCND3
|
cyclin D3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF740_ZNF219
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0060032 | notochord regression(GO:0060032) |
| 1.1 | 3.2 | GO:1904828 | regulation of phenotypic switching by transcription from RNA polymerase II promoter(GO:0100057) regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.8 | 3.8 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.7 | 6.3 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.6 | 1.9 | GO:0097086 | amniotic stem cell differentiation(GO:0097086) negative regulation of dense core granule biogenesis(GO:2000706) negative regulation of mesenchymal stem cell differentiation(GO:2000740) regulation of amniotic stem cell differentiation(GO:2000797) negative regulation of amniotic stem cell differentiation(GO:2000798) |
| 0.6 | 1.9 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.6 | 7.9 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.5 | 1.5 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.5 | 1.9 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.5 | 1.4 | GO:0060302 | negative regulation of cytokine activity(GO:0060302) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.5 | 1.4 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.5 | 2.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.4 | 2.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.4 | 2.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.4 | 5.0 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.4 | 2.8 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.4 | 3.6 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.4 | 1.9 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.4 | 1.1 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.4 | 1.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.4 | 0.7 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.4 | 1.4 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.3 | 1.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.3 | 1.4 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.3 | 0.7 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.3 | 1.3 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.3 | 1.3 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.3 | 2.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 4.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.3 | 1.5 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.3 | 5.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.3 | 4.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.3 | 3.8 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.3 | 0.6 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.3 | 1.4 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.3 | 1.7 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.3 | 1.6 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.3 | 0.5 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.3 | 4.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.3 | 0.8 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.3 | 0.8 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.3 | 0.8 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.3 | 1.8 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 3.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 1.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 | 1.5 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.2 | 0.7 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 1.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 4.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.2 | 1.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 0.7 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 1.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.2 | GO:0042779 | tRNA 3'-trailer cleavage, endonucleolytic(GO:0034414) tRNA 3'-trailer cleavage(GO:0042779) |
| 0.2 | 1.4 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.2 | 2.7 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 0.5 | GO:0072366 | regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.2 | 2.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 1.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 1.9 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 | 3.6 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.2 | 4.0 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 2.3 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.2 | 2.4 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.2 | 0.6 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.2 | 0.6 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 3.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 0.8 | GO:0032904 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.2 | 1.4 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) response to fluoride(GO:1902617) |
| 0.2 | 1.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 1.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 1.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.2 | 0.6 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.2 | 3.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 0.6 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.2 | 0.6 | GO:1902811 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.2 | 0.6 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.2 | 1.3 | GO:2000771 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.2 | 1.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 1.4 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.2 | 0.7 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 | 3.1 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.2 | 0.9 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 1.0 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.2 | 0.5 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.2 | 0.5 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.2 | 0.9 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.2 | 1.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 2.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.2 | 1.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.2 | 0.2 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.2 | 0.5 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.2 | 2.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 1.0 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.2 | 0.5 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.1 | 1.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.6 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.1 | 1.0 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.4 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.5 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.8 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 1.4 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.7 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.9 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.5 | GO:2000595 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.1 | 0.6 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 2.6 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.5 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 1.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.5 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 7.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.8 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 0.3 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 0.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 1.6 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.1 | 1.6 | GO:0051382 | centromere complex assembly(GO:0034508) kinetochore assembly(GO:0051382) |
| 0.1 | 0.4 | GO:0032597 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.1 | 0.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 3.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 1.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 2.3 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.1 | 0.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 1.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.4 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.4 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.7 | GO:0071317 | cellular response to morphine(GO:0071315) cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.1 | 0.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 3.0 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 5.9 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.3 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 0.3 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.1 | 1.8 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 2.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 19.6 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.1 | 1.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.8 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.1 | 0.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 1.8 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 1.2 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 2.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.3 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.6 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.1 | 0.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.5 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.2 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.1 | 1.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 0.4 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 0.9 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.8 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.1 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 8.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.3 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 2.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.3 | GO:0070428 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.1 | 0.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.7 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.8 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 1.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 2.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.6 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 1.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.4 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 1.0 | GO:0007512 | adult heart development(GO:0007512) |
| 0.1 | 0.8 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 | 3.2 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 0.4 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 1.9 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 2.2 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 1.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 1.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 3.9 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.2 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.6 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 1.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 2.1 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 1.9 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.1 | 1.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 3.2 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 1.0 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.7 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.2 | GO:0090467 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 1.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 2.4 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 1.2 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.5 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 1.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.8 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.2 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.5 | GO:0071492 | response to UV-A(GO:0070141) cellular response to UV-A(GO:0071492) |
| 0.0 | 0.8 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.6 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.3 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 5.9 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.1 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.8 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 0.2 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 1.0 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.6 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.3 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.3 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 2.0 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.3 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 3.1 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 0.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.9 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.3 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:2000382 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 3.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.4 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.8 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 2.9 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 1.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 1.2 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 4.1 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.3 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 1.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 1.7 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.7 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 2.0 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 1.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.4 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 1.9 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 1.1 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.5 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.1 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.3 | GO:0021924 | cell proliferation in hindbrain(GO:0021534) cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 | 1.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 4.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.3 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.4 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 | 0.5 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 1.0 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.4 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0051796 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 1.3 | GO:0003279 | cardiac septum development(GO:0003279) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 1.1 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.1 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.0 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.5 | 2.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.5 | 1.9 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.4 | 1.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.4 | 1.3 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.4 | 2.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.4 | 3.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 1.1 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.3 | 6.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.3 | 3.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.3 | 7.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.3 | 13.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 0.8 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.3 | 0.8 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 1.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.2 | 5.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 3.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 0.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 3.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 2.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 3.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 3.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 2.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 1.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 3.0 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 1.0 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.5 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 0.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 1.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 3.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.7 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.1 | 2.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 3.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 12.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 7.8 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 2.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 1.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.5 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 1.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 5.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.3 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 1.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 2.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 1.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 3.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 4.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 4.7 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 0.8 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 1.2 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 4.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.0 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 3.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.5 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.4 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 2.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 4.2 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.8 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 11.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 4.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 3.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 3.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 2.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 4.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 3.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 79.3 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 1.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 1.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 10.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.5 | 1.5 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.5 | 4.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.4 | 1.3 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.4 | 3.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 3.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.3 | 2.6 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.3 | 7.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 2.7 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.3 | 7.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 0.8 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.3 | 1.8 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 6.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 15.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 0.7 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.2 | 1.2 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.2 | 1.3 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 6.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 0.6 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.2 | 1.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 1.7 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 0.6 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 0.2 | 1.6 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.2 | 7.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 5.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 1.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 3.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 4.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 3.6 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 1.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.2 | 0.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 4.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 9.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 1.4 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.0 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 1.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 1.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.5 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 1.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 1.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 3.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 1.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 14.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 4.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.4 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 4.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 3.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.9 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 4.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 3.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 4.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.9 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 0.5 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 4.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.8 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 1.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.5 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 2.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.4 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 1.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 3.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 3.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 2.3 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 1.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 1.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 3.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 3.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 10.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.5 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.1 | 0.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 3.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 4.7 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 17.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 0.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.3 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 1.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.3 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 3.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.7 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 2.9 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0019808 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.8 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 2.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 8.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 3.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 23.5 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 3.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 4.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 2.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 3.9 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 4.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 2.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 1.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 25.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.8 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 4.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 14.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 7.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 4.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 5.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 3.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 4.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 2.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 5.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 7.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 4.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.5 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 3.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 4.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 0.4 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 6.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 7.9 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.1 | 3.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 5.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 6.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 4.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 3.7 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 2.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.5 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.1 | 2.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 1.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 3.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 4.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 2.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 4.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 2.1 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 1.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.8 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 13.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 3.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 2.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.6 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |